Idealized Sensitivity Study
Contents
Idealized Sensitivity Study¶
Throughout this document unmodified is unsed to describe the runs which used the original values of LANDUSE.TBL.
import pandas as pd
import numpy as np
import xarray as xr
from glob import glob
import matplotlib.pyplot as plt
import numpy as np
from datetime import datetime, timedelta
import matplotlib.pyplot as plt
from matplotlib.cm import get_cmap
from matplotlib.dates import DateFormatter
from netCDF4 import Dataset
from wrf import (getvar, to_np, vertcross, smooth2d, CoordPair,
get_basemap, latlon_coords, g_geoht, combine_files, ALL_TIMES)
# Suppress warnings so the notebook looks nice
import warnings
warnings.filterwarnings('ignore')
# Style for plotting
plt.style.use('default')
# N-ICE Measurements
## NOTE: SEB Measurements are in UTC
### Importing SEB dataset
Measurements_seb = xr.open_dataset('/Users/smurphy/all_datasets/nice_published_datasets/N-ICE_sebData_v1.nc', decode_times = False)
### Longwave radiation into dataframes
M_downlw = pd.DataFrame(Measurements_seb.variables['surface_downwelling_longwave_flux'].values,
index = pd.to_datetime(Measurements_seb.variables['unix_time'].values, unit = 's'), columns = ['lw'])
M_uplw = pd.DataFrame(Measurements_seb.variables['surface_upwelling_longwave_flux'].values,
index = pd.to_datetime(Measurements_seb.variables['unix_time'].values, unit = 's'), columns = ['lw'])
### Shortwave radiation into dataframes
M_downsw = pd.DataFrame(Measurements_seb.variables['surface_downwelling_shortwave_flux'].values,
index = pd.to_datetime(Measurements_seb.variables['unix_time'].values, unit = 's'), columns = ['sw'])
M_upsw = pd.DataFrame(Measurements_seb.variables['surface_upwelling_shortwave_flux'].values,
index = pd.to_datetime(Measurements_seb.variables['unix_time'].values, unit = 's'), columns = ['sw'])
### Calculating net radiation
M_net = (M_downlw['lw'] - M_uplw['lw']) + (M_downsw['sw'] - M_upsw['sw'])
### Sensible and latent heat flux into dataframes
M_lat = -pd.DataFrame(Measurements_seb.variables['surface_downward_latent_heat_flux'].values,
index = pd.to_datetime(Measurements_seb.variables['unix_time'].values, unit = 's'), columns = ['lh'])
M_sen = -pd.DataFrame(Measurements_seb.variables['surface_downward_sensible_heat_flux'].values,
index = pd.to_datetime(Measurements_seb.variables['unix_time'].values, unit = 's'), columns = ['sh'])
## NOTE: sensible and latent heat flux negative to match WRF sign convention
### Temperature
T_meas = pd.read_excel('/Users/smurphy/Documents/PhD/datasets/nice_data/Ts.xlsx', index_col = 0)
## Soundings
MeasSoundings = xr.open_dataset('/Users/smurphy/all_datasets/nice_published_datasets/rsData_gridded.nc')
MeasSoundings_Times = pd.read_fwf('/Users/smurphy/all_datasets/nice_published_datasets/rsData_gridded_dates.txt', header = None)
MeasSoundings_Times.columns = ['year','month','day','hour','minute','second']
MeasSoundings_Times.index = pd.to_datetime(MeasSoundings_Times[['year', 'month', 'day', 'hour', 'minute', 'second']])
sounding_t = pd.DataFrame(MeasSoundings['temp'].values, index = MeasSoundings_Times.index, columns = MeasSoundings['height'].values)
sounding_ws = pd.DataFrame(MeasSoundings['ws'].values, index = MeasSoundings_Times.index, columns = MeasSoundings['height'].values)
sfc_relative_humidity = pd.DataFrame(MeasSoundings['rh'].values,
index = MeasSoundings_Times.index,
columns = MeasSoundings['height'].astype(str))
sfc_wind_direction = pd.DataFrame(MeasSoundings['wd'].values,
index = MeasSoundings_Times.index,
columns = MeasSoundings['height'].astype(str))
sfc_wind_speed = pd.DataFrame(MeasSoundings['ws'].values,
index = MeasSoundings_Times.index,
columns = MeasSoundings['height'].astype(str))
sfc_temperature = pd.DataFrame(MeasSoundings['temp'].values + 273.15,
index = MeasSoundings_Times.index,
columns = MeasSoundings['height'].astype(str))
sfc_pressure = pd.DataFrame(MeasSoundings['press'].values,
index = MeasSoundings_Times.index,
columns = MeasSoundings['height'].astype(str))
# Height above sea level (m)
sfc_height = sfc_relative_humidity.index.values
# Potential temperature (k)
k = 2/7 # constent for potential temperature equation
sfc_potential_temperature = sfc_temperature * (1000 / sfc_pressure) ** k
# Mixing ratio (g/kg)
# Using Clasius Clapperyon
# Saturation vapor pressure #hPa
sfc_saturation_vapor_pressure = 6.112 * np.exp(((17.67 * (sfc_temperature - 273.15)) / ((sfc_temperature - 273.15) + 243.5)))
# Vapor pressure
sfc_vapor_pressure = sfc_saturation_vapor_pressure * (sfc_relative_humidity / 100)
# Mixing ratio g/kg
sfc_mixing_ratio = 621.97 * (sfc_vapor_pressure / ((sfc_pressure) - sfc_vapor_pressure))
sfc_mixing_ratio.index = sfc_mixing_ratio.index.to_pydatetime()
# Set date format for plots throughout the notebook
myFmt = DateFormatter("%m/%d \n %H:%M:%S")
Case 1 - Winter Clear¶
fns = glob('/Volumes/seagate_desktop/idealized/case1/000101/wrfo*')
wrflist = list()
for fn in fns:
wrflist.append(Dataset(fn))
# Defining start and end dates of case study
sdate = '2015-02-04'
edate = '2015-02-06'
sebmask = (M_downlw.index > sdate) & (M_downlw.index < edate)
#cldfra = getvar(wrflist, "CLDFRA", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
z = getvar(wrflist, "z").mean('south_north').mean('west_east')
#cldfra_df = pd.DataFrame(cldfra.values, index = cldfra.Time.values, columns = z)
lh = getvar(wrflist, "LH", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lh_df = pd.DataFrame(lh.values, index = lh.Time.values)
sh = getvar(wrflist, "HFX", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
sh_df = pd.DataFrame(sh.values, index = sh.Time.values)
lwdnb = getvar(wrflist, "LWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwdnb_df = pd.DataFrame(lwdnb.values, index = lwdnb.Time.values)
lwupb = getvar(wrflist, "LWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwupb_df = pd.DataFrame(lwupb.values, index = lwupb.Time.values)
swdnb = getvar(wrflist, "SWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swdnb_df = pd.DataFrame(swdnb.values, index = swdnb.Time.values)
swupb = getvar(wrflist, "SWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swupb_df = pd.DataFrame(swupb.values, index = swupb.Time.values)
# Finding all wrfout file
fns = glob('/Volumes/seagate_desktop/idealized/landusetbl_modifications/1ClearWinter_000101/wrfo*')
# Creating an empty list to append to
wrflist = list()
# Opening the wrfout files and appending them to the empty list
for fn in fns:
wrflist.append(Dataset(fn))
#cldfra_mod = getvar(wrflist, "CLDFRA", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
z_mod = getvar(wrflist, "z").mean('south_north').mean('west_east')
#cldfra_df_mod = pd.DataFrame(cldfra_mod.values, index = cldfra_mod.Time.values, columns = z_mod)
## lh - latent heat flux
lh_mod = getvar(wrflist, "LH", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lh_df_mod = pd.DataFrame(lh_mod.values, index = lh_mod.Time.values)
## sh - sensible heat flux
sh_mod = getvar(wrflist, "HFX", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
sh_df_mod = pd.DataFrame(sh_mod.values, index = sh_mod.Time.values)
## lwdnb - downwelling longwave radiation
lwdnb_mod = getvar(wrflist, "LWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwdnb_df_mod = pd.DataFrame(lwdnb_mod.values, index = lwdnb_mod.Time.values)
## lwupb - upwelling longwave radiation
lwupb_mod = getvar(wrflist, "LWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwupb_df_mod = pd.DataFrame(lwupb_mod.values, index = lwupb_mod.Time.values)
## swdnb - downwelling shortwave radiation
swdnb_mod = getvar(wrflist, "SWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swdnb_df_mod = pd.DataFrame(swdnb_mod.values, index = swdnb_mod.Time.values)
## swupb - upwelling shortwave radiation
swupb_mod = getvar(wrflist, "SWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swupb_df_mod = pd.DataFrame(swupb_mod.values, index = swupb_mod.Time.values)
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
/var/folders/21/bhksj_cn7zz43y8x0m4gsh0h0000gn/T/ipykernel_54832/1427607582.py in <module>
67
68 ## swupb - upwelling shortwave radiation
---> 69 swupb_mod = getvar(wrflist, "SWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
70 swupb_df_mod = pd.DataFrame(swupb_mod.values, index = swupb_mod.Time.values)
~/opt/anaconda3/lib/python3.8/site-packages/wrf/routines.py in getvar(wrfin, varname, timeidx, method, squeeze, cache, meta, **kwargs)
334 if is_standard_wrf_var(wrfin, varname) and varname != "Times":
335 _check_kargs("default", kwargs)
--> 336 return extract_vars(wrfin, timeidx, varname,
337 method, squeeze, cache, meta, _key)[varname]
338 elif varname == "Times":
~/opt/anaconda3/lib/python3.8/site-packages/wrf/util.py in extract_vars(wrfin, timeidx, varnames, method, squeeze, cache, meta, _key)
2276 varlist = varnames
2277
-> 2278 return {var:_extract_var(wrfin, var, timeidx, None,
2279 method, squeeze, cache, meta, _key)
2280 for var in varlist}
~/opt/anaconda3/lib/python3.8/site-packages/wrf/util.py in <dictcomp>(.0)
2276 varlist = varnames
2277
-> 2278 return {var:_extract_var(wrfin, var, timeidx, None,
2279 method, squeeze, cache, meta, _key)
2280 for var in varlist}
~/opt/anaconda3/lib/python3.8/site-packages/wrf/util.py in _extract_var(wrfin, varname, timeidx, is_moving, method, squeeze, cache, meta, _key)
2213 else:
2214 # Squeeze handled in this routine, so just return it
-> 2215 return combine_files(wrfin, varname, timeidx, is_moving,
2216 method, squeeze, meta, _key)
2217
~/opt/anaconda3/lib/python3.8/site-packages/wrf/util.py in combine_files(wrfin, varname, timeidx, is_moving, method, squeeze, meta, _key)
2115 outarr = _combine_dict(wrfseq, varname, timeidx, method, meta, _key)
2116 elif method.lower() == "cat":
-> 2117 outarr = _cat_files(wrfseq, varname, timeidx, is_moving,
2118 squeeze, meta, _key)
2119 elif method.lower() == "join":
~/opt/anaconda3/lib/python3.8/site-packages/wrf/util.py in _cat_files(wrfseq, varname, timeidx, is_moving, squeeze, meta, _key)
1685 break
1686 else:
-> 1687 vardata = wrfnc.variables[varname][:]
1688
1689 numtimes = vardata.shape[0]
netCDF4/_netCDF4.pyx in netCDF4._netCDF4.Variable.__getitem__()
netCDF4/_netCDF4.pyx in netCDF4._netCDF4.Variable._toma()
~/opt/anaconda3/lib/python3.8/site-packages/numpy/__init__.py in __getattr__(attr)
267 # Warn for expired attributes, and return a dummy function
268 # that always raises an exception.
--> 269 try:
270 msg = __expired_functions__[attr]
271 except KeyError:
KeyboardInterrupt:
wrfstat = xr.open_dataset('/Volumes/seagate_desktop/idealized/case1/000101/wrfstat_d01_2015-02-04_00:00:00')
cst_qc = wrfstat['CSP_QC']
cst_tsk = wrfstat['CST_TSK']
cst_sh = wrfstat['CST_SH']
cst_lh = wrfstat['CST_LH']
cst_time = wrfstat['Times']
csp_z = wrfstat['CSP_Z']
csp_u = wrfstat['CSP_U']
csp_v = wrfstat['CSP_V']
csv_qv = wrfstat["CSP_QV"]
qc = pd.DataFrame(cst_qc.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
sh_df = pd.DataFrame(cst_sh.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'), columns = ['sh'])
lh_df = pd.DataFrame(cst_lh.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'), columns = ['lh'])
tsk_df = pd.DataFrame(cst_tsk.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'), columns = ['tsk'])
u_df = pd.DataFrame(csp_u.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
v_df = pd.DataFrame(csp_v.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
ws_df = np.sqrt(v_df**2 + u_df**2)
qv_df = pd.DataFrame(csv_qv.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
cloudmask = wrfstat['CSV_CLDFRAC'].isel(south_north = round(len(wrfstat.south_north) / 2), west_east = round(len(wrfstat.west_east) / 2))
cloudmask_df = pd.DataFrame(cloudmask.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 22:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
wrfstat = xr.open_dataset('/Volumes/seagate_desktop/idealized/landusetbl_modifications/1ClearWinter_000101/wrfstat_d01_2015-02-04_00:00:00')
cst_qc = wrfstat['CSP_QC']
cst_tsk = wrfstat['CST_TSK']
cst_sh = wrfstat['CST_SH']
cst_lh = wrfstat['CST_LH']
cst_time = wrfstat['Times']
csp_z = wrfstat['CSP_Z']
csp_u = wrfstat['CSP_U']
csp_v = wrfstat['CSP_V']
csv_qv = wrfstat["CSP_QV"]
qc_mod = pd.DataFrame(cst_qc.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
sh_df_mod = pd.DataFrame(cst_sh.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'), columns = ['sh'])
lh_df_mod = pd.DataFrame(cst_lh.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'), columns = ['lh'])
tsk_df_mod = pd.DataFrame(cst_tsk.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'), columns = ['tsk'])
u_df_mod = pd.DataFrame(csp_u.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
v_df_mod = pd.DataFrame(csp_v.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
ws_df_mod = np.sqrt(v_df_mod**2 + u_df_mod**2)
qv_df_mod = pd.DataFrame(csv_qv.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
cloudmask = wrfstat['CSV_CLDFRAC'].isel(south_north = round(len(wrfstat.south_north) / 2), west_east = round(len(wrfstat.west_east) / 2))
cloudmask_df_mod = pd.DataFrame(cloudmask.values,
index = pd.date_range(start='2/4/2015 00:00:00', end='2/6/2015 21:00:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
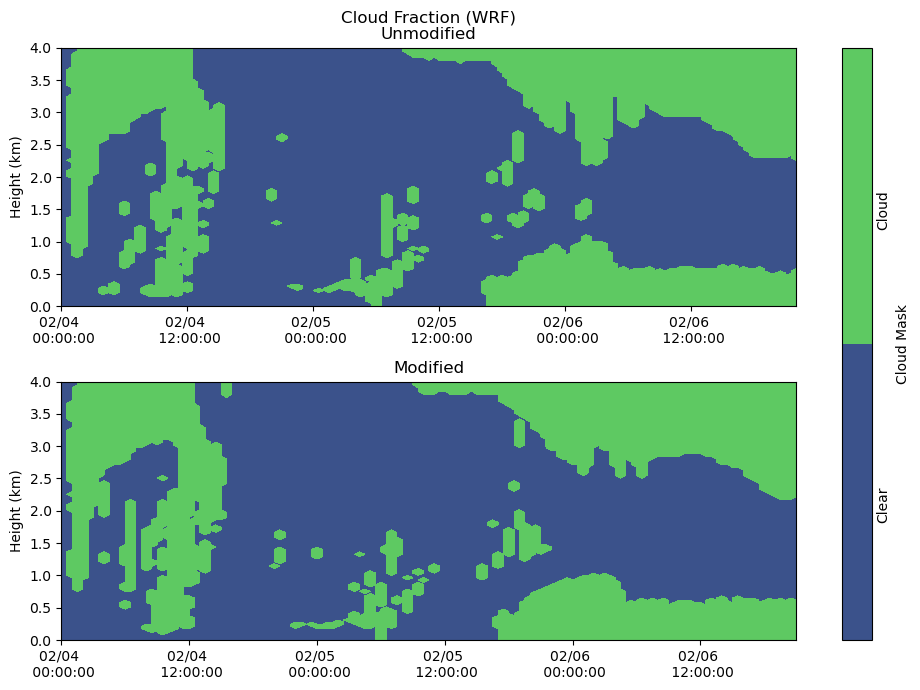
Clouds¶
fig, axs = plt.subplots(2, figsize=(10,7))
heatmap = axs[0].contourf(cloudmask_df.index, cloudmask_df.columns / 1000, cloudmask_df.T.values, levels = [-1, 0, 1])
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Cloud Fraction (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,4)
heatmap = axs[1].contourf(cloudmask_df_mod.index, cloudmask_df_mod.columns / 1000, cloudmask_df_mod.T, levels = [-1, 0, 1])
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,4)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[:], ticks = [-0.5, 0.5], label = "Cloud Mask")
cbar.ax.set_yticklabels(['Clear','Cloud'], rotation = 90)
cbar.ax.tick_params(size=0)
plt.show()
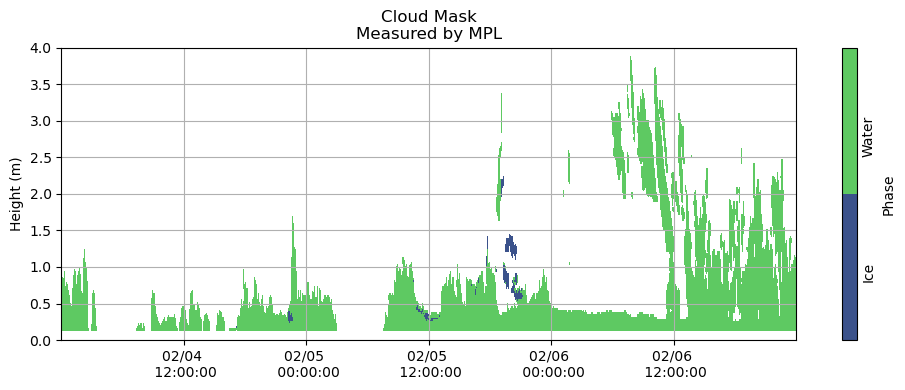
fns = ['/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150204.cdf',
'/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150205.cdf',
'/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150206.cdf']
measured_cloudmask_all = pd.DataFrame()
for fn in fns:
rbt_cldmask = xr.open_dataset(fn)
# Time is in UTC time stamp in fractional hours
Time = rbt_cldmask.variables['DataTime'].values
# Fix time to be a timestamp - Currently in hours
yy = 2015
mm = int(fn[-8:-6])
dd = int(fn[-6:-4])
date = [datetime(yy,mm,dd) + timedelta(seconds=hr*3600) for hr in Time[~np.isnan(Time)]]
measured_cloudmask = pd.DataFrame(rbt_cldmask['PhaseMask'][~np.isnan(Time)].values, index = date, columns = rbt_cldmask['Range'])
measured_cloudmask_all = pd.concat([measured_cloudmask_all, measured_cloudmask])
fig, ax = plt.subplots(figsize=(10,4))
plt.grid()
heatmap = plt.contourf(measured_cloudmask_all.index,
measured_cloudmask_all.columns / 1000,
measured_cloudmask_all.T.values,
[0, 1, 2])
# nan - no information
# 1 - cloud liquid
# 2 - cloud ice
plt.title('Cloud Mask\nMeasured by MPL')
plt.ylabel('Height (m)')
cbar = fig.colorbar(heatmap, label = 'Phase', ticks = [0.5, 1.5], orientation = 'vertical')
cbar.ax.set_yticklabels(['Ice', 'Water'], rotation = 90)
cbar.ax.tick_params(size=0)
plt.gca().xaxis.set_major_formatter(myFmt)
plt.ylim(0, 4)
plt.tight_layout()
plt.show()
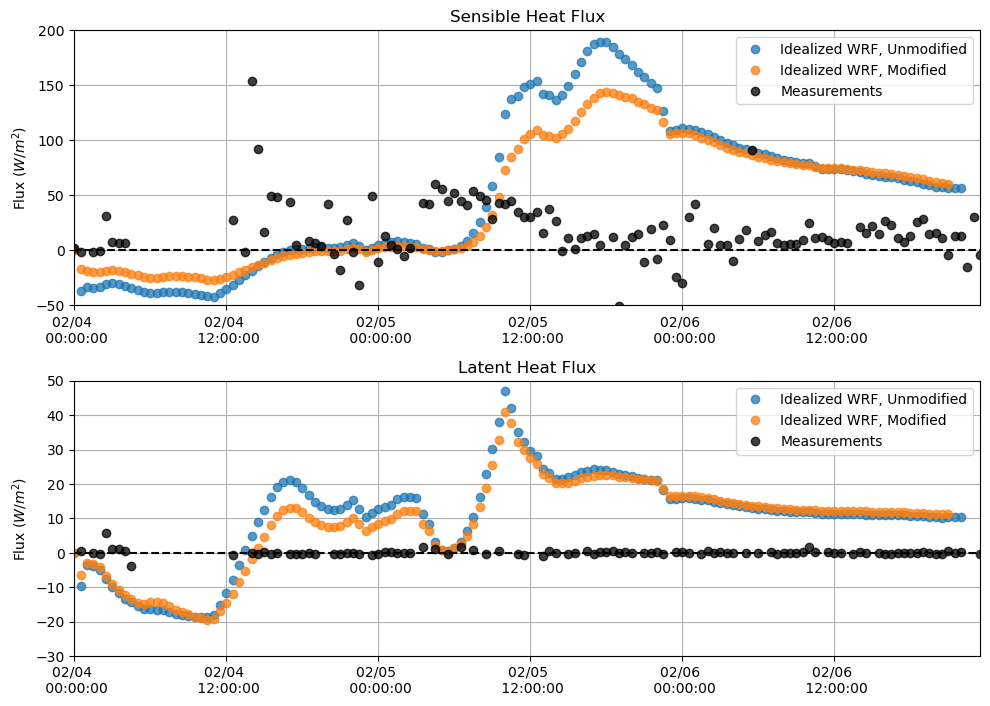
Sensible and Latent Heat Flux¶
plt.figure(figsize = (10,7))
plt.subplot(211)
plt.plot(sh_df, 'o', alpha = 0.75)
plt.plot(sh_df_mod, 'o', alpha = 0.75)
plt.plot(M_sen[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.hlines(0, xmin = M_lat[sdate:edate].index[0], xmax = M_lat[sdate:edate].index[-1], linestyle = '--', color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Sensible Heat Flux')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(-50,200)
plt.xlim(M_lat[sdate:edate].index[0],M_lat[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(212)
plt.plot(lh_df, 'o', alpha = 0.75)
plt.plot(lh_df_mod, 'o', alpha = 0.75)
plt.plot(M_lat[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.hlines(0, xmin = M_lat[sdate:edate].index[0], xmax = M_lat[sdate:edate].index[-1], linestyle = '--', color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Latent Heat Flux')
plt.grid()
plt.ylim(-30,50)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_lat[sdate:edate].index[0],M_lat[sdate:edate].index[-1])
plt.tight_layout()
plt.gca().xaxis.set_major_formatter(myFmt)
plt.show()
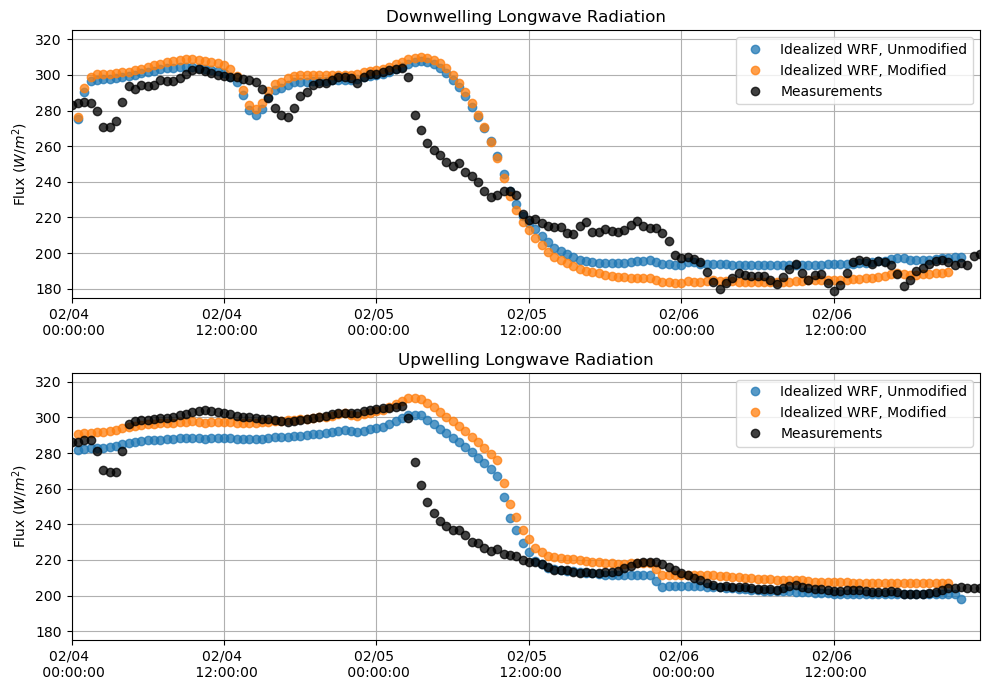
Longwave Radiation¶
plt.figure(figsize = (10,7))
plt.subplot(211)
plt.plot(lwdnb_df, 'o', alpha = 0.75)
plt.plot(lwdnb_df_mod, 'o', alpha = 0.75)
plt.plot(M_downlw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Downwelling Longwave Radiation')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(175,325)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(212)
plt.plot(lwupb_df, 'o', alpha = 0.75)
plt.plot(lwupb_df_mod, 'o', alpha = 0.75)
plt.plot(M_uplw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Upwelling Longwave Radiation')
plt.grid()
plt.ylim(175,325)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.tight_layout()
plt.show()
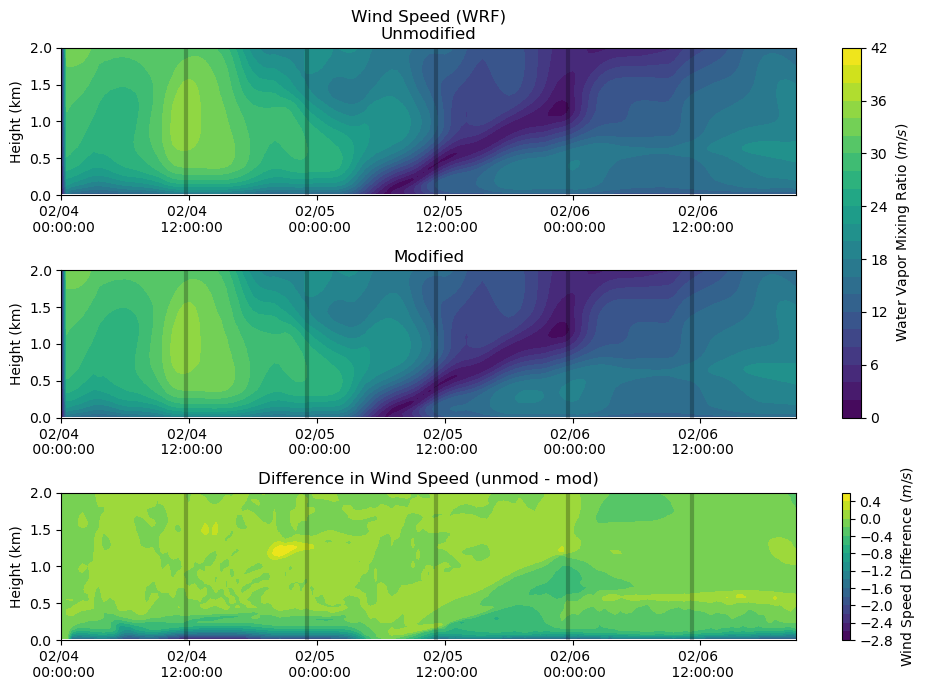
Wind Speed¶
fig, axs = plt.subplots(3, figsize=(10,7))
heatmap = axs[0].contourf(ws_df_mod.index, ws_df.columns / 1000, ws_df.T.values[:,:-2], levels = 20)
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Wind Speed (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,2)
axs[0].vlines(sounding_ws[sdate:'2015-02-06'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap = axs[1].contourf(ws_df_mod.index, ws_df_mod.columns / 1000, ws_df_mod.T.values, levels = 20)
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,2)
axs[1].vlines(sounding_ws[sdate:'2015-02-06'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap_1 = axs[2].contourf(ws_df_mod.index, ws_df.columns / 1000, ws_df.T.values[:,:-2] - ws_df_mod.T.values, levels = 20)
axs[2].set_ylabel('Height (km)')
axs[2].set_title('Difference in Wind Speed (unmod - mod)')
axs[2].xaxis.set_major_formatter(myFmt)
axs[2].set_ylim(0,2)
axs[2].vlines(sounding_ws[sdate:'2015-02-06'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[0:2])
cbar.set_label('Water Vapor Mixing Ratio ($m/s$)')
cbar = fig.colorbar(heatmap_1, ax=axs[2])
cbar.set_label('Wind Speed Difference ($m/s$)')
plt.show()
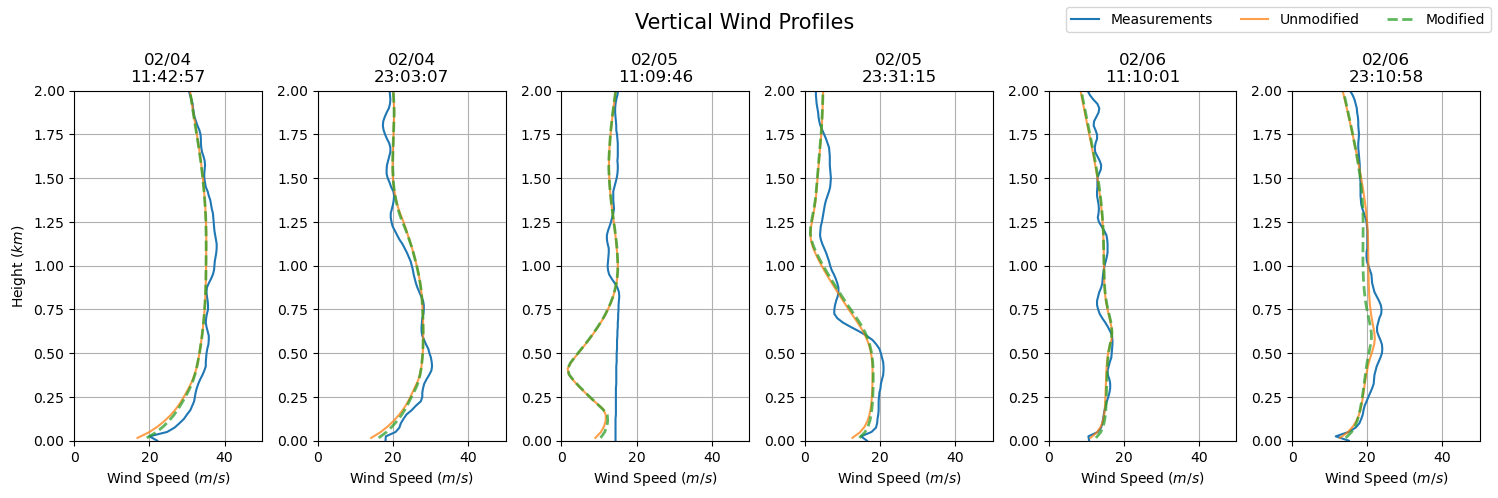
dates = sounding_ws[sdate:'2015-02-06'].index
fig, axs = plt.subplots(ncols = len(dates), figsize=(15,5))
for i in np.arange(0, len(dates), 1):
prof = axs[i].plot(sounding_ws.loc[dates[i]], sounding_ws.loc[dates[i]].index / 1000, label = 'Measurements')
axs[i].set_ylim(0, 2)
axs[i].grid()
axs[i].set_xlim(0, 50)
axs[i].set_xlabel('Wind Speed ($m/s$)')
axs[i].set_title(dates[i].strftime('%m/%d') + '\n' + dates[i].strftime('%H:%M:%S'))
axs[i].plot(ws_df.iloc[ws_df.index.get_loc(dates[i], method = 'nearest')], ws_df.iloc[ws_df.index.get_loc(dates[0], method = 'nearest')].index / 1000, label = 'Unmodified', alpha = 0.75)
axs[i].plot(ws_df_mod.iloc[ws_df_mod.index.get_loc(dates[i], method = 'nearest')], ws_df_mod.iloc[ws_df_mod.index.get_loc(dates[0], method = 'nearest')].index / 1000, '--', label = 'Modified', alpha = 0.75, lw = 2)
axs[0].set_ylabel('Height ($km$)')
plt.suptitle('Vertical Wind Profiles', fontsize = 15)
plt.tight_layout()
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right', ncol = 3)
plt.show()
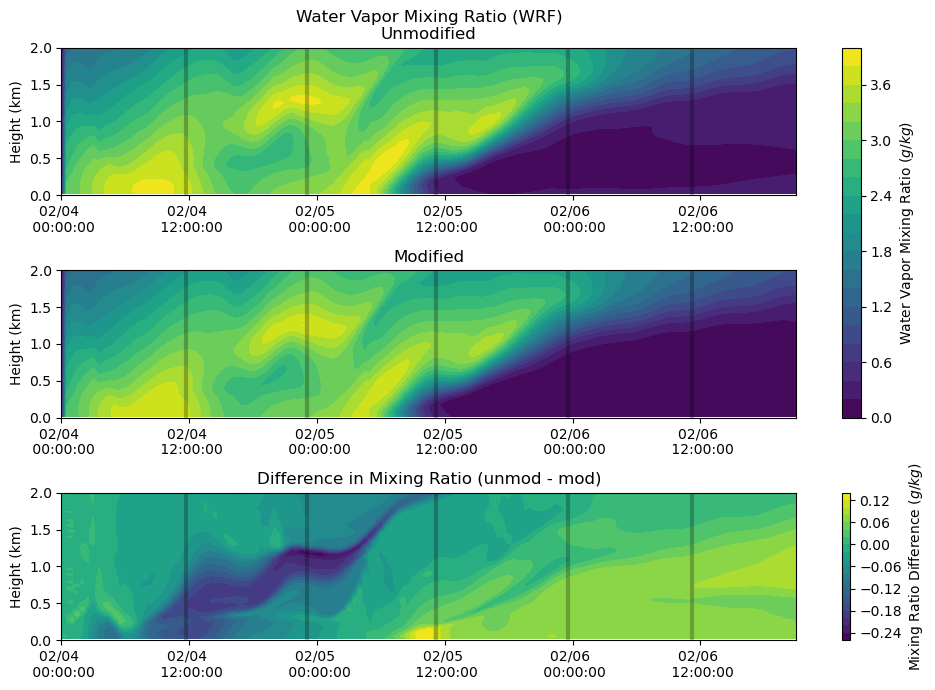
Moisture¶
fig, axs = plt.subplots(3, figsize=(10,7))
heatmap = axs[0].contourf(qv_df_mod.index, qv_df.columns / 1000, qv_df.T.values[:,:-2] * 1000, levels = 20)
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Water Vapor Mixing Ratio (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,2)
axs[0].vlines(sounding_ws[sdate:'2015-02-06'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap = axs[1].contourf(qv_df_mod.index, qv_df_mod.columns / 1000, qv_df_mod.T.values * 1000, levels = 20)
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,2)
axs[1].vlines(sounding_ws[sdate:'2015-02-06'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap_1 = axs[2].contourf(qv_df_mod.index, qv_df.columns / 1000,
(qv_df.T.values[:,:-2] * 1000) - (qv_df_mod.T.values * 1000), levels = 20)
axs[2].set_ylabel('Height (km)')
axs[2].set_title('Difference in Mixing Ratio (unmod - mod)')
axs[2].xaxis.set_major_formatter(myFmt)
axs[2].set_ylim(0,2)
axs[2].vlines(sounding_ws[sdate:'2015-02-06'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[0:2])
cbar.set_label('Water Vapor Mixing Ratio ($g/kg$)')
cbar = fig.colorbar(heatmap_1, ax=axs[2])
cbar.set_label('Mixing Ratio Difference ($g/kg$)')
plt.show()
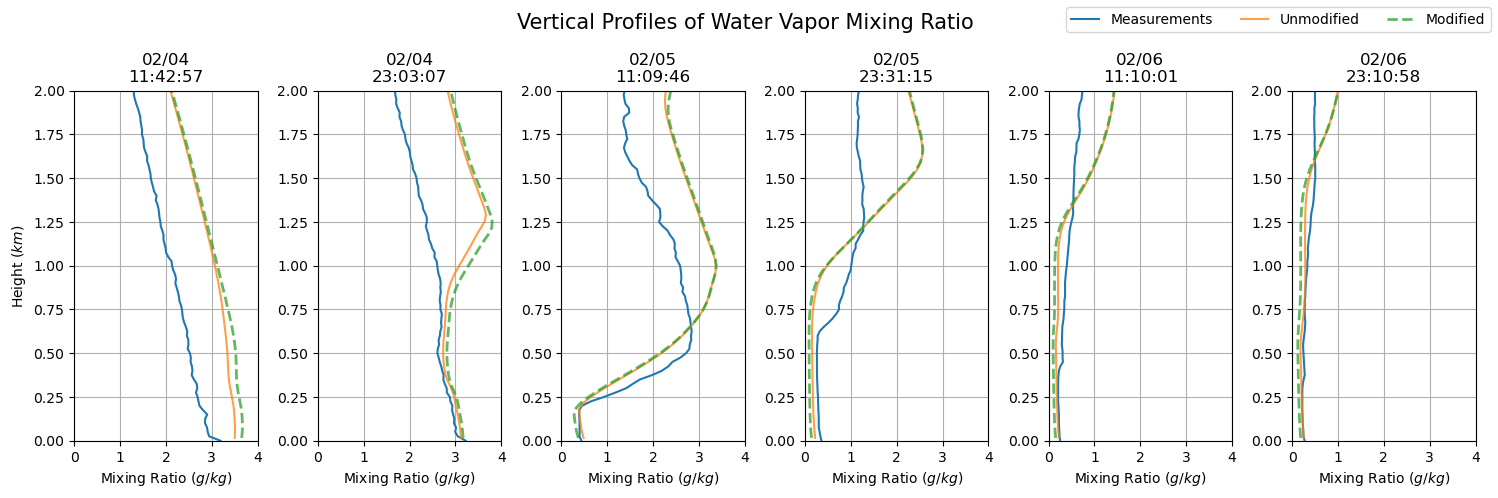
dates = sfc_mixing_ratio[sdate:'2015-02-06'].index
fig, axs = plt.subplots(ncols = len(dates), figsize=(15,5))
for i in np.arange(0, len(dates), 1):
prof = axs[i].plot(sfc_mixing_ratio.loc[dates[i].to_pydatetime()],
sfc_mixing_ratio.loc[dates[i].to_pydatetime()].index.astype(float) / 1000,
label = 'Measurements')
axs[i].set_ylim(0, 2)
axs[i].grid()
axs[i].set_xlim(0, 4)
axs[i].set_xlabel('Mixing Ratio ($g/kg$)')
axs[i].set_title(dates[i].strftime('%m/%d') + '\n' + dates[i].strftime('%H:%M:%S'))
axs[i].plot(qv_df.iloc[qv_df.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')] * 1000,
qv_df.iloc[qv_df.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')].index / 1000,
label = 'Unmodified', alpha = 0.75)
axs[i].plot(qv_df_mod.iloc[qv_df_mod.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')] * 1000,
qv_df_mod.iloc[qv_df_mod.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')].index / 1000,
'--', label = 'Modified', alpha = 0.75, lw = 2)
axs[0].set_ylabel('Height ($km$)')
plt.suptitle('Vertical Profiles of Water Vapor Mixing Ratio', fontsize = 15)
plt.tight_layout()
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right', ncol = 3)
plt.show()
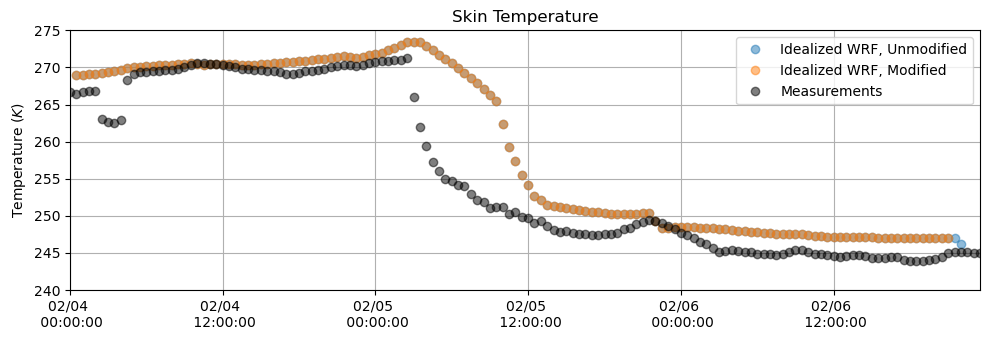
Skin Temperature¶
plt.figure(figsize = (10,3.5))
plt.plot(tsk_df, 'o', alpha = 0.5)
plt.plot(tsk_df_mod, 'o', alpha = 0.5)
plt.plot(T_meas[sdate:edate].resample('30min').first(), 'o', alpha = 0.5, color = 'k')
plt.ylabel('Temperature $(K)$')
plt.title('Skin Temperature')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(240,275)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.tight_layout()
plt.show()
Summary Table¶
Red highlighting indicates the lowest correlation of that variable, green indicates the highest.
edate = '2015-02-06 21:00:00'
lwdns = M_downlw[sdate:edate]
lwdns.columns = ['measured']
lwdns['unmodified'] = lwdnb_df[sdate:edate]
lwdns['modified'] = lwdnb_df_mod[sdate:edate]
lwups = M_uplw[sdate:edate]
lwups.columns = ['measured']
lwups['unmodified'] = lwupb_df[sdate:edate]
lwups['modified'] = lwupb_df_mod[sdate:edate]
swdns = M_downsw[sdate:edate]
swdns.columns = ['measured']
swdns['unmodified'] = swdnb_df[sdate:edate]
swdns['modified'] = swdnb_df_mod[sdate:edate]
swups = M_upsw[sdate:edate]
swups.columns = ['measured']
swups['unmodified'] = swupb_df[sdate:edate]
swups['modified'] = swupb_df_mod[sdate:edate]
lhs = M_lat[sdate:edate]
lhs.columns = ['measured']
lhs['unmodified'] = lh_df[sdate:edate]
lhs['modified'] = lh_df_mod[sdate:edate]
shs = M_sen[sdate:edate]
shs.columns = ['measured']
shs['unmodified'] = sh_df[sdate:edate]
shs['modified'] = sh_df_mod[sdate:edate]
correlation_coefficients = pd.DataFrame([lhs.corr()['measured'].values,
shs.corr()['measured'].values,
lwdns.corr()['measured'].values,
lwups.corr()['measured'].values],
columns = lhs.corr().columns,
index = ['Latent', 'Sensible', 'Downwelling Longwave', 'Upwelling Longwave']).T
cc_1 = correlation_coefficients
r_squared = correlation_coefficients ** 2
rs_1 = r_squared
r_squared[1:].style.highlight_max(color = 'lightgreen', axis = 0).highlight_min(color = 'pink', axis = 0)
| Latent | Sensible | Downwelling Longwave | Upwelling Longwave | |
|---|---|---|---|---|
| unmodified | 0.003053 | 0.031668 | 0.712964 | 0.609335 |
| modified | 0.000586 | 0.051439 | 0.744958 | 0.609335 |
Case 2 - Spring Cloudy¶
fns = glob('/Volumes/seagate_desktop/idealized/case4/000101/wrfo*')
wrflist = list()
for fn in fns:
wrflist.append(Dataset(fn))
sdate = '2015-05-02'
edate = '2015-05-04'
sebmask = (M_downlw.index > sdate) & (M_downlw.index < edate)
#cldfra = getvar(wrflist, "CLDFRA", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
z = getvar(wrflist, "z").mean('south_north').mean('west_east')
#cldfra_df = pd.DataFrame(cldfra.values, index = cldfra.Time.values, columns = z)
lh = getvar(wrflist, "LH", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lh_df = pd.DataFrame(lh.values, index = lh.Time.values)
sh = getvar(wrflist, "HFX", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
sh_df = pd.DataFrame(sh.values, index = sh.Time.values)
lwdnb = getvar(wrflist, "LWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwdnb_df = pd.DataFrame(lwdnb.values, index = lwdnb.Time.values)
lwupb = getvar(wrflist, "LWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwupb_df = pd.DataFrame(lwupb.values, index = lwupb.Time.values)
swdnb = getvar(wrflist, "SWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swdnb_df = pd.DataFrame(swdnb.values, index = swdnb.Time.values)
swupb = getvar(wrflist, "SWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swupb_df = pd.DataFrame(swupb.values, index = swupb.Time.values)
# Finding all wrfout file
fns = glob('/Volumes/seagate_desktop/idealized/landusetbl_modifications/2CloudySpring_000101/wrfo*')
# Creating an empty list to append to
wrflist = list()
# Opening the wrfout files and appending them to the empty list
for fn in fns:
wrflist.append(Dataset(fn))
#cldfra_mod = getvar(wrflist, "CLDFRA", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
z_mod = getvar(wrflist, "z").mean('south_north').mean('west_east')
#cldfra_df_mod = pd.DataFrame(cldfra_mod.values, index = cldfra_mod.Time.values, columns = z_mod)
## lh - latent heat flux
lh_mod = getvar(wrflist, "LH", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lh_df_mod = pd.DataFrame(lh_mod.values, index = lh_mod.Time.values)
## sh - sensible heat flux
sh_mod = getvar(wrflist, "HFX", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
sh_df_mod = pd.DataFrame(sh_mod.values, index = sh_mod.Time.values)
## lwdnb - downwelling longwave radiation
lwdnb_mod = getvar(wrflist, "LWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwdnb_df_mod = pd.DataFrame(lwdnb_mod.values, index = lwdnb_mod.Time.values)
## lwupb - upwelling longwave radiation
lwupb_mod = getvar(wrflist, "LWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwupb_df_mod = pd.DataFrame(lwupb_mod.values, index = lwupb_mod.Time.values)
## swdnb - downwelling shortwave radiation
swdnb_mod = getvar(wrflist, "SWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swdnb_df_mod = pd.DataFrame(swdnb_mod.values, index = swdnb_mod.Time.values)
## swupb - upwelling shortwave radiation
swupb_mod = getvar(wrflist, "SWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swupb_df_mod = pd.DataFrame(swupb_mod.values, index = swupb_mod.Time.values)
wrfstat = xr.open_dataset('/Volumes/seagate_desktop/idealized/case4/000101/wrfstat_d01_2015-05-02_00:00:00')
cst_qc = wrfstat['CSP_QC']
cst_tsk = wrfstat['CST_TSK']
cst_sh = wrfstat['CST_SH']
cst_lh = wrfstat['CST_LH']
cst_time = wrfstat['Times']
csp_z = wrfstat['CSP_Z']
csp_u = wrfstat['CSP_U']
csp_v = wrfstat['CSP_V']
csv_qv = wrfstat["CSP_QV"]
qc = pd.DataFrame(cst_qc.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
sh_df = pd.DataFrame(cst_sh.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'), columns = ['sh'])
lh_df = pd.DataFrame(cst_lh.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'), columns = ['lh'])
tsk_df = pd.DataFrame(cst_tsk.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'), columns = ['tsk'])
u_df = pd.DataFrame(csp_u.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
v_df = pd.DataFrame(csp_v.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
ws_df = np.sqrt(v_df**2 + u_df**2)
qv_df = pd.DataFrame(csv_qv.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
cloudmask = wrfstat['CSV_CLDFRAC'].isel(south_north = round(len(wrfstat.south_north) / 2), west_east = round(len(wrfstat.west_east) / 2))
cloudmask_df = pd.DataFrame(cloudmask.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 19:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
wrfstat = xr.open_dataset('/Volumes/seagate_desktop/idealized/landusetbl_modifications/2CloudySpring_000101/wrfstat_d01_2015-05-02_00:00:00')
cst_qc = wrfstat['CSP_QC']
cst_tsk = wrfstat['CST_TSK']
cst_sh = wrfstat['CST_SH']
cst_lh = wrfstat['CST_LH']
cst_time = wrfstat['Times']
csp_z = wrfstat['CSP_Z']
csp_u = wrfstat['CSP_U']
csp_v = wrfstat['CSP_V']
csv_qv = wrfstat["CSP_QV"]
qc_mod = pd.DataFrame(cst_qc.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
sh_df_mod = pd.DataFrame(cst_sh.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'), columns = ['sh'])
lh_df_mod = pd.DataFrame(cst_lh.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'), columns = ['lh'])
tsk_df_mod = pd.DataFrame(cst_tsk.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'), columns = ['tsk'])
u_df_mod = pd.DataFrame(csp_u.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
v_df_mod = pd.DataFrame(csp_v.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
ws_df_mod = np.sqrt(v_df_mod**2 + u_df_mod**2)
qv_df_mod = pd.DataFrame(csv_qv.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
cloudmask = wrfstat['CSV_CLDFRAC'].isel(south_north = round(len(wrfstat.south_north) / 2), west_east = round(len(wrfstat.west_east) / 2))
cloudmask_df_mod = pd.DataFrame(cloudmask.values,
index = pd.date_range(start='5/2/2015 00:00:00', end='5/4/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
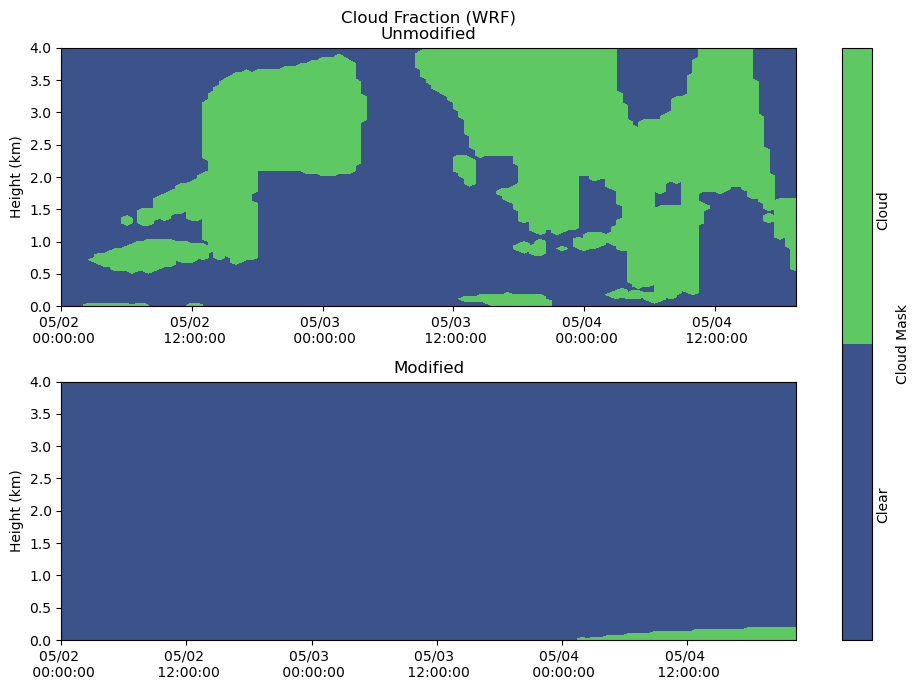
Clouds¶
fig, axs = plt.subplots(2, figsize=(10,7))
heatmap = axs[0].contourf(cloudmask_df.index, cloudmask_df.columns / 1000, cloudmask_df.T.values, levels = [-1, 0, 1])
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Cloud Fraction (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,4)
heatmap = axs[1].contourf(cloudmask_df_mod.index, cloudmask_df_mod.columns / 1000, cloudmask_df_mod.T, levels = [-1, 0, 1])
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,4)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[:], ticks = [-0.5, 0.5], label = "Cloud Mask")
cbar.ax.set_yticklabels(['Clear','Cloud'], rotation = 90)
cbar.ax.tick_params(size=0)
plt.show()
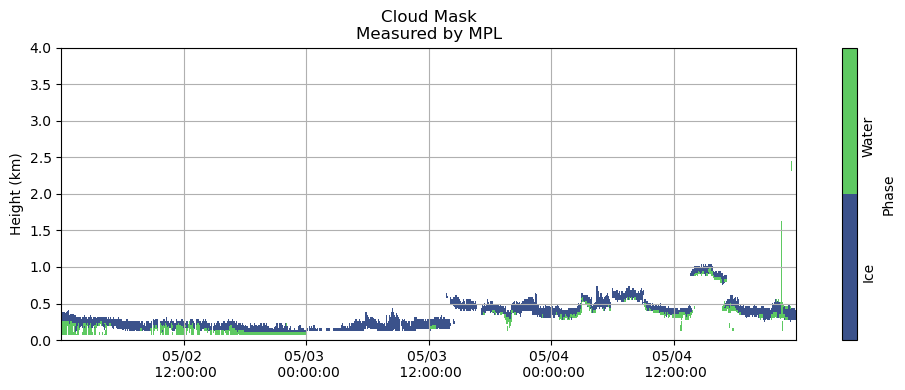
fns = ['/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150502.cdf',
'/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150503.cdf',
'/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150504.cdf']
measured_cloudmask_all = pd.DataFrame()
for fn in fns:
rbt_cldmask = xr.open_dataset(fn)
# Time is in UTC time stamp in fractional hours
Time = rbt_cldmask.variables['DataTime'].values
# Fix time to be a timestamp - Currently in hours
yy = 2015
mm = int(fn[-8:-6])
dd = int(fn[-6:-4])
date = [datetime(yy,mm,dd) + timedelta(seconds=hr*3600) for hr in Time[~np.isnan(Time)]]
measured_cloudmask = pd.DataFrame(rbt_cldmask['PhaseMask'][~np.isnan(Time)].values, index = date, columns = rbt_cldmask['Range'])
measured_cloudmask_all = pd.concat([measured_cloudmask_all, measured_cloudmask])
fig, ax = plt.subplots(figsize=(10,4))
plt.grid()
heatmap = plt.contourf(measured_cloudmask_all.index,
measured_cloudmask_all.columns / 1000,
measured_cloudmask_all.T.values,
[0, 1, 2])
# nan - no information
# 1 - cloud liquid
# 2 - cloud ice
plt.title('Cloud Mask\nMeasured by MPL')
plt.ylabel('Height (km)')
cbar = fig.colorbar(heatmap, label = 'Phase', ticks = [0.5, 1.5], orientation = 'vertical')
cbar.ax.set_yticklabels(['Ice', 'Water'], rotation = 90)
cbar.ax.tick_params(size=0)
plt.gca().xaxis.set_major_formatter(myFmt)
plt.ylim(0, 4)
plt.tight_layout()
plt.show()
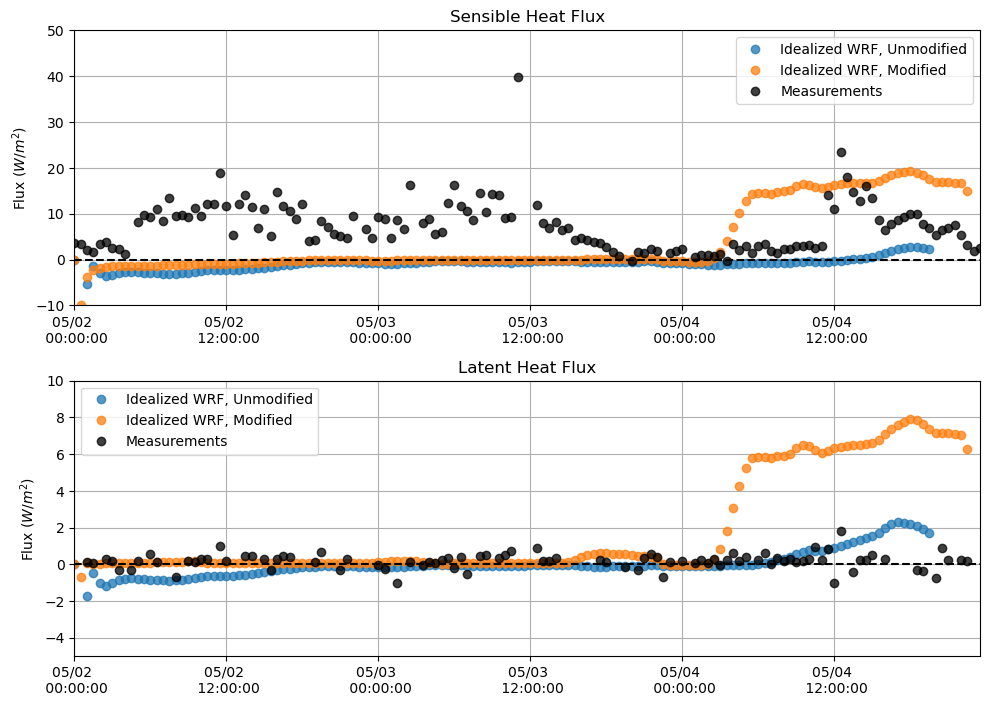
Sensible and Latent Heat Flux¶
plt.figure(figsize = (10,7))
plt.subplot(211)
plt.plot(sh_df, 'o', alpha = 0.75)
plt.plot(sh_df_mod, 'o', alpha = 0.75)
plt.plot(M_sen[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.hlines(0, xmin = M_lat[sdate:edate].index[0], xmax = M_lat[sdate:edate].index[-1], linestyle = '--', color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Sensible Heat Flux')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(-10,50)
plt.xlim(M_lat[sdate:edate].index[0],M_lat[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(212)
plt.plot(lh_df, 'o', alpha = 0.75)
plt.plot(lh_df_mod, 'o', alpha = 0.75)
plt.plot(M_lat[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.hlines(0, xmin = M_lat[sdate:edate].index[0], xmax = M_lat[sdate:edate].index[-1], linestyle = '--', color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Latent Heat Flux')
plt.grid()
plt.ylim(-5,10)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_lat[sdate:edate].index[0],M_lat[sdate:edate].index[-1])
plt.tight_layout()
plt.gca().xaxis.set_major_formatter(myFmt)
plt.show()
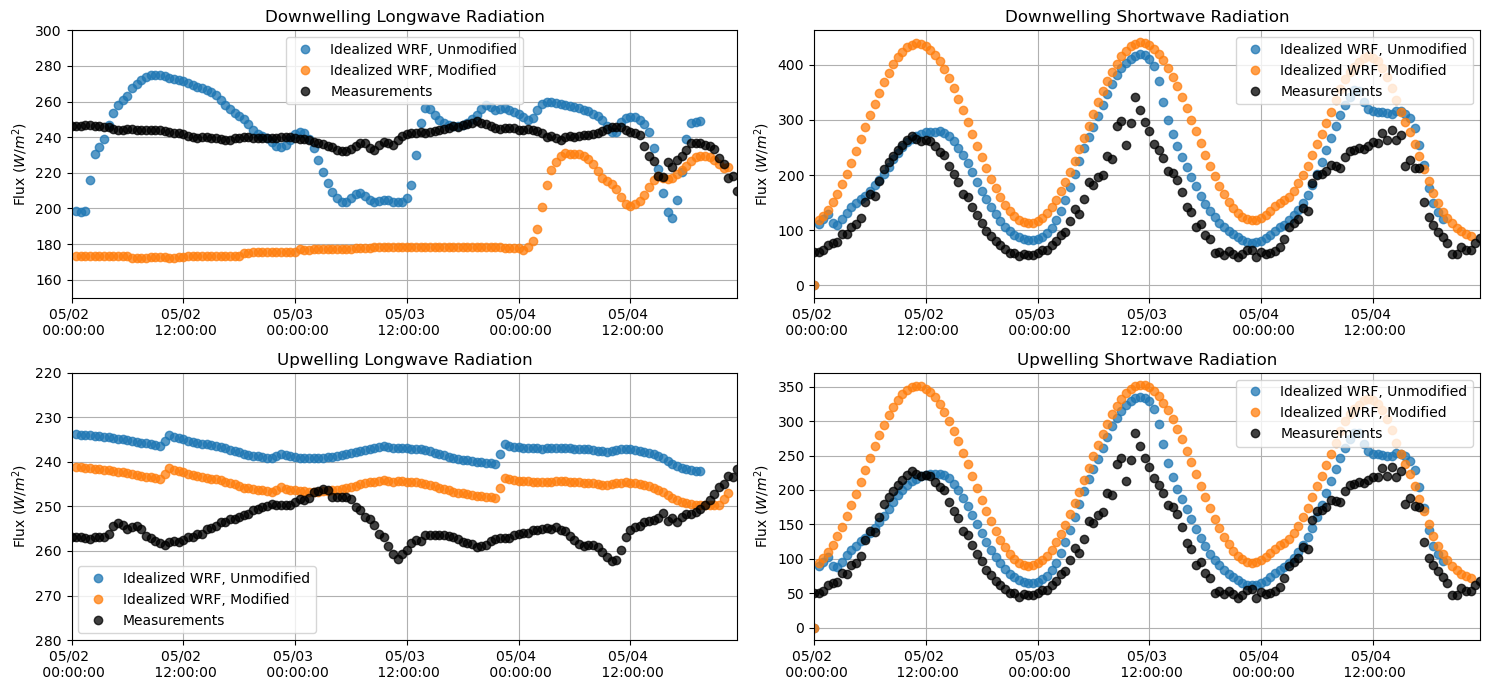
Longwave and Shortwave Radiation¶
plt.figure(figsize = (15,7))
plt.subplot(221)
plt.plot(lwdnb_df, 'o', alpha = 0.75)
plt.plot(lwdnb_df_mod, 'o', alpha = 0.75)
plt.plot(M_downlw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Downwelling Longwave Radiation')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(150,300)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(223)
plt.plot(lwupb_df, 'o', alpha = 0.75)
plt.plot(lwupb_df_mod, 'o', alpha = 0.75)
plt.plot(M_uplw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Upwelling Longwave Radiation')
plt.grid()
plt.ylim(280,220)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(222)
plt.plot(swdnb_df, 'o', alpha = 0.75)
plt.plot(swdnb_df_mod, 'o', alpha = 0.75)
plt.plot(M_downsw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Downwelling Shortwave Radiation')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
#plt.ylim(180,300)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(224)
plt.plot(swupb_df, 'o', alpha = 0.75)
plt.plot(swupb_df_mod, 'o', alpha = 0.75)
plt.plot(M_upsw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Upwelling Shortwave Radiation')
plt.grid()
#plt.ylim(180, 300)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.tight_layout()
plt.show()
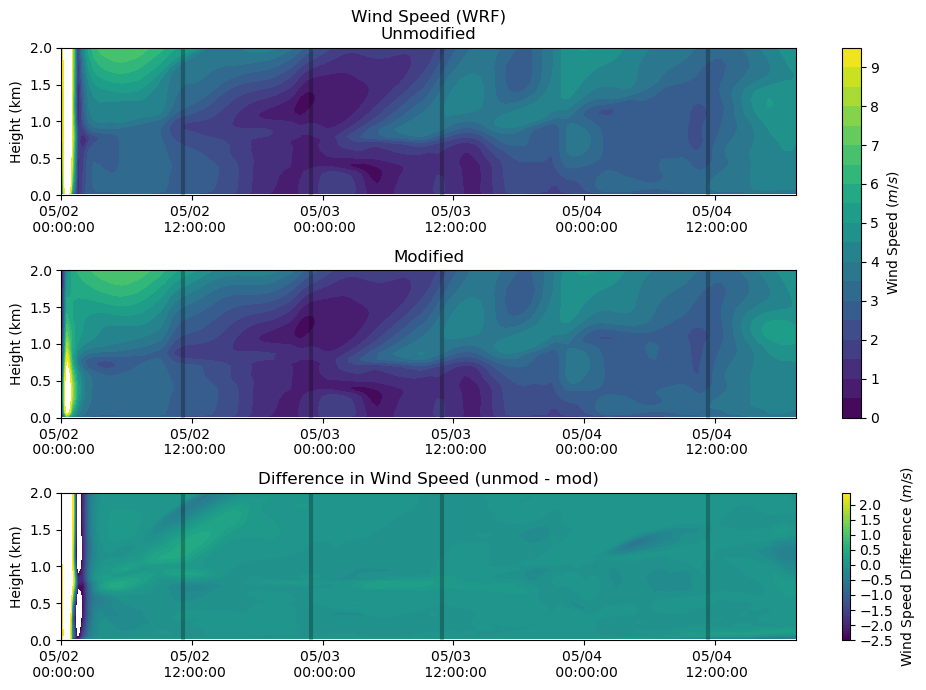
Wind Speed¶
fig, axs = plt.subplots(3, figsize=(10,7))
levels = np.arange(0,10,0.5)
heatmap = axs[0].contourf(ws_df.index, ws_df.columns / 1000, ws_df.T.values, levels = levels)
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Wind Speed (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,2)
axs[0].vlines(sounding_ws[sdate:'2015-05-04'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap = axs[1].contourf(ws_df.index, ws_df_mod.columns / 1000, ws_df_mod.T.values[:,:-6], levels = levels)
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,2)
axs[1].vlines(sounding_ws[sdate:'2015-05-04'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
levels = np.arange(-2.5,2.5,0.1)
heatmap_1 = axs[2].contourf(ws_df.index, ws_df.columns / 1000, ws_df.T.values - ws_df_mod.T.values[:,:-6], levels = levels)
axs[2].set_ylabel('Height (km)')
axs[2].set_title('Difference in Wind Speed (unmod - mod)')
axs[2].xaxis.set_major_formatter(myFmt)
axs[2].set_ylim(0,2)
axs[2].vlines(sounding_ws[sdate:'2015-05-04'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[0:2])
cbar.set_label('Wind Speed ($m/s$)')
cbar = fig.colorbar(heatmap_1, ax=axs[2])
cbar.set_label('Wind Speed Difference ($m/s$)')
plt.show()
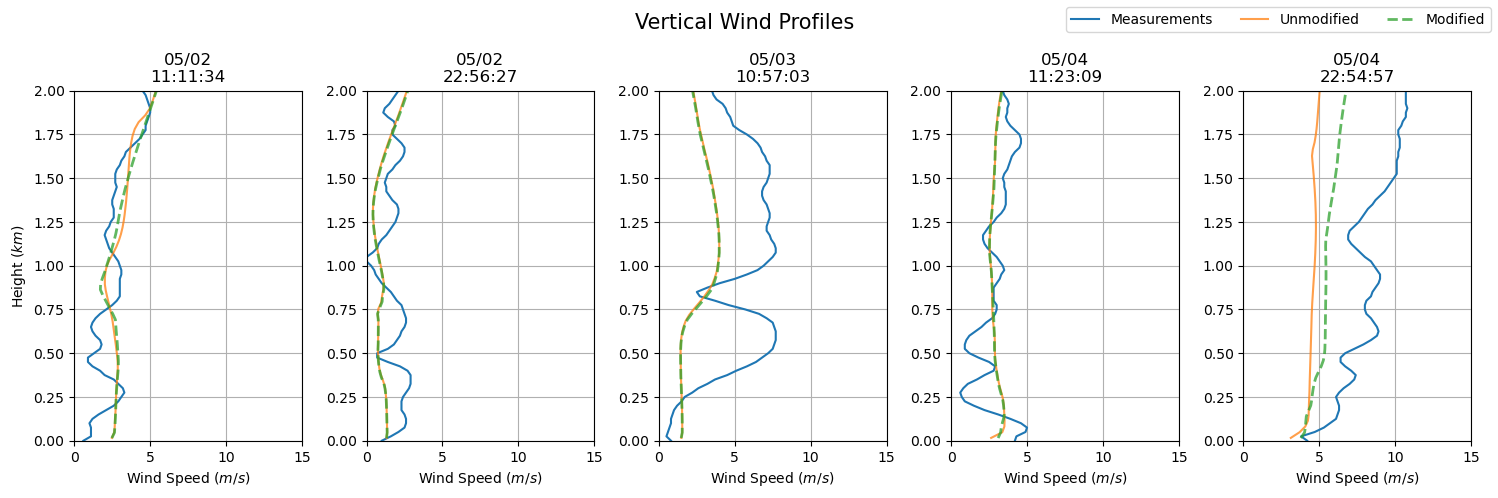
dates = sounding_ws[sdate:'2015-05-04'].index
fig, axs = plt.subplots(ncols = len(dates), figsize=(15,5))
for i in np.arange(0, len(dates), 1):
prof = axs[i].plot(sounding_ws.loc[dates[i]], sounding_ws.loc[dates[i]].index / 1000, label = 'Measurements')
axs[i].set_ylim(0, 2)
axs[i].grid()
axs[i].set_xlim(0, 15)
axs[i].set_xlabel('Wind Speed ($m/s$)')
axs[i].set_title(dates[i].strftime('%m/%d') + '\n' + dates[i].strftime('%H:%M:%S'))
axs[i].plot(ws_df.iloc[ws_df.index.get_loc(dates[i], method = 'nearest')], ws_df.iloc[ws_df.index.get_loc(dates[0], method = 'nearest')].index / 1000, label = 'Unmodified', alpha = 0.75)
axs[i].plot(ws_df_mod.iloc[ws_df_mod.index.get_loc(dates[i], method = 'nearest')], ws_df_mod.iloc[ws_df_mod.index.get_loc(dates[0], method = 'nearest')].index / 1000, '--', label = 'Modified', alpha = 0.75, lw = 2)
axs[0].set_ylabel('Height ($km$)')
plt.suptitle('Vertical Wind Profiles', fontsize = 15)
plt.tight_layout()
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right', ncol = 3)
plt.show()
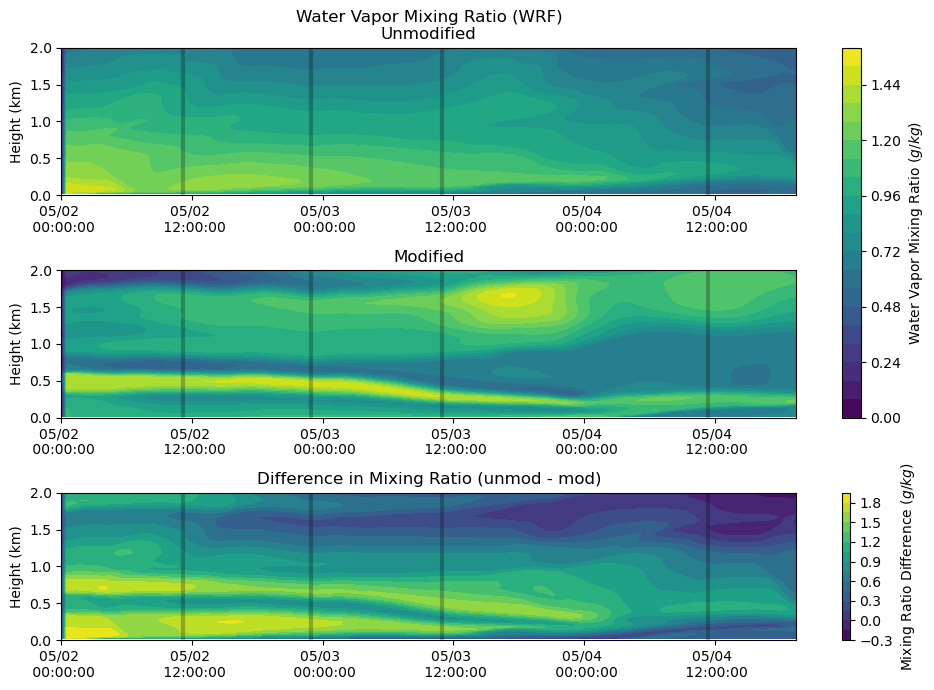
Moisture¶
fig, axs = plt.subplots(3, figsize=(10,7))
heatmap = axs[0].contourf(qv_df.index, qv_df.columns / 1000, qv_df.T.values * 1000, levels = 20)
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Water Vapor Mixing Ratio (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,2)
axs[0].vlines(sounding_ws[sdate:'2015-05-04'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap = axs[1].contourf(qv_df.index, qv_df_mod.columns / 1000, qv_df_mod.T.values[:,:-6] * 1000, levels = 20)
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,2)
axs[1].vlines(sounding_ws[sdate:'2015-05-04'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap_1 = axs[2].contourf(qv_df.index, qv_df.columns / 1000,
(qv_df.T.values * 1000) - (qv_df_mod.T.values[:,:-6] * 1000), levels = 20)
axs[2].set_ylabel('Height (km)')
axs[2].set_title('Difference in Mixing Ratio (unmod - mod)')
axs[2].xaxis.set_major_formatter(myFmt)
axs[2].set_ylim(0,2)
axs[2].vlines(sounding_ws[sdate:'2015-05-04'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[0:2])
cbar.set_label('Water Vapor Mixing Ratio ($g/kg$)')
cbar = fig.colorbar(heatmap_1, ax=axs[2])
cbar.set_label('Mixing Ratio Difference ($g/kg$)')
plt.show()
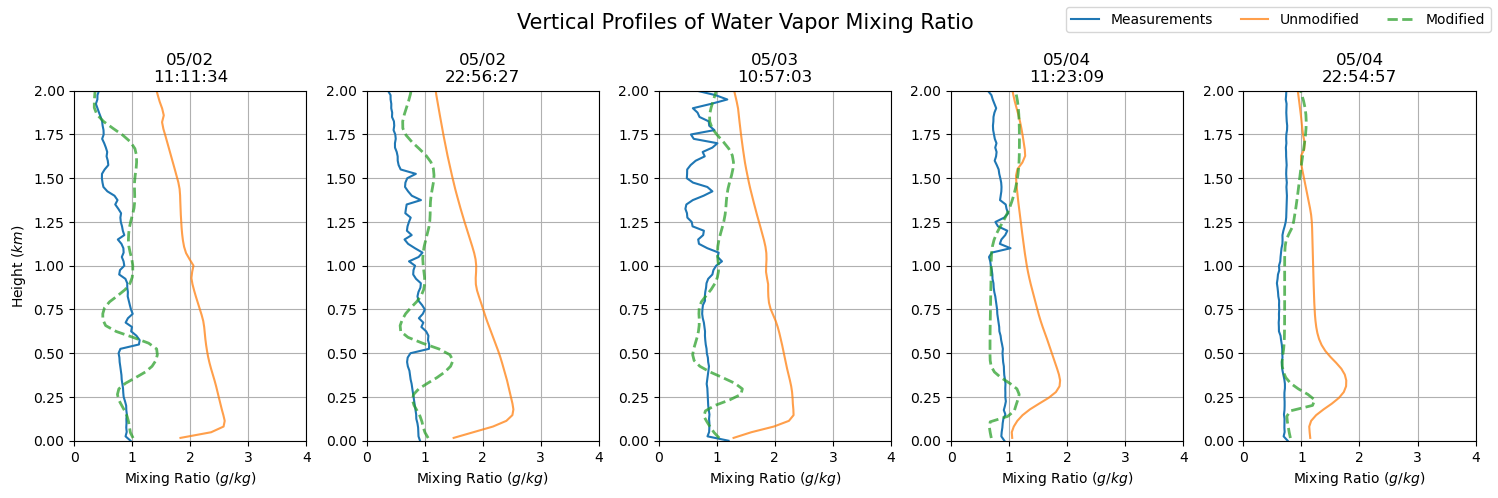
dates = sfc_mixing_ratio[sdate:'2015-05-04'].index
fig, axs = plt.subplots(ncols = len(dates), figsize=(15,5))
for i in np.arange(0, len(dates), 1):
prof = axs[i].plot(sfc_mixing_ratio.loc[dates[i].to_pydatetime()],
sfc_mixing_ratio.loc[dates[i].to_pydatetime()].index.astype(float) / 1000,
label = 'Measurements')
axs[i].set_ylim(0, 2)
axs[i].grid()
axs[i].set_xlim(0, 4)
axs[i].set_xlabel('Mixing Ratio ($g/kg$)')
axs[i].set_title(dates[i].strftime('%m/%d') + '\n' + dates[i].strftime('%H:%M:%S'))
axs[i].plot(qv_df.iloc[qv_df.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')] * 1000,
qv_df.iloc[qv_df.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')].index / 1000,
label = 'Unmodified', alpha = 0.75)
axs[i].plot(qv_df_mod.iloc[qv_df_mod.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')] * 1000,
qv_df_mod.iloc[qv_df_mod.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')].index / 1000,
'--', label = 'Modified', alpha = 0.75, lw = 2)
axs[0].set_ylabel('Height ($km$)')
plt.suptitle('Vertical Profiles of Water Vapor Mixing Ratio', fontsize = 15)
plt.tight_layout()
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right', ncol = 3)
plt.show()
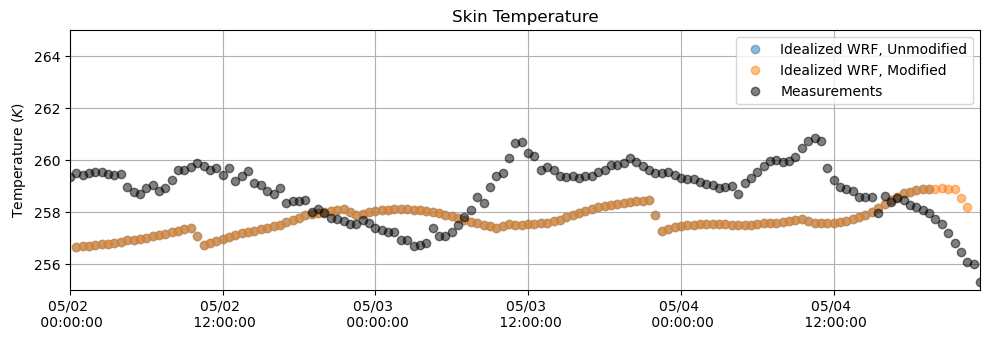
Skin Temperature¶
plt.figure(figsize = (10,3.5))
plt.plot(tsk_df, 'o', alpha = 0.5)
plt.plot(tsk_df_mod, 'o', alpha = 0.5)
plt.plot(T_meas[sdate:edate].resample('30min').first(), 'o', alpha = 0.5, color = 'k')
plt.ylabel('Temperature $(K)$')
plt.title('Skin Temperature')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(255,265)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.tight_layout()
plt.show()
Summary Table¶
Red highlighting indicates the lowest correlation of that variable, green indicates the highest.
edate = '2015-05-04 19:30:00'
lwdns = M_downlw[sdate:edate]
lwdns.columns = ['measured']
lwdns['unmodified'] = lwdnb_df[sdate:edate]
lwdns['modified'] = lwdnb_df_mod[sdate:edate]
lwups = M_uplw[sdate:edate]
lwups.columns = ['measured']
lwups['unmodified'] = lwupb_df[sdate:edate]
lwups['modified'] = lwupb_df_mod[sdate:edate]
swdns = M_downsw[sdate:edate]
swdns.columns = ['measured']
swdns['unmodified'] = swdnb_df[sdate:edate]
swdns['modified'] = swdnb_df_mod[sdate:edate]
swups = M_upsw[sdate:edate]
swups.columns = ['measured']
swups['unmodified'] = swupb_df[sdate:edate]
swups['modified'] = swupb_df_mod[sdate:edate]
lhs = M_lat[sdate:edate]
lhs.columns = ['measured']
lhs['unmodified'] = lh_df[sdate:edate]
lhs['modified'] = lh_df_mod[sdate:edate]
shs = M_sen[sdate:edate]
shs.columns = ['measured']
shs['unmodified'] = sh_df[sdate:edate]
shs['modified'] = sh_df_mod[sdate:edate]
correlation_coefficients = pd.DataFrame([lhs.corr()['measured'].values,
shs.corr()['measured'].values,
lwdns.corr()['measured'].values,
lwups.corr()['measured'].values,
swdns.corr()['measured'].values,
swups.corr()['measured'].values],
columns = lhs.corr().columns,
index = ['Latent', 'Sensible', 'Downwelling Longwave', 'Upwelling Longwave', 'Downwelling Shortwave', 'Upwelling Shortwave']).T
cc_2 = correlation_coefficients
r_squared = correlation_coefficients ** 2
rs_2 = r_squared
r_squared[1:].style.highlight_max(color = 'lightgreen', axis = 0).highlight_min(color = 'pink', axis = 0)
| Latent | Sensible | Downwelling Longwave | Upwelling Longwave | Downwelling Shortwave | Upwelling Shortwave | |
|---|---|---|---|---|---|---|
| unmodified | 0.000645 | 0.004846 | 0.091075 | 0.006432 | 0.885284 | 0.883314 |
| modified | 0.007810 | 0.001159 | 0.108260 | 0.006431 | 0.886173 | 0.895837 |
Case 3 - Spring Clear¶
sdate = '2015-05-22'
edate = '2015-05-24'
M_net = (M_downlw['lw'] - M_uplw['lw'])
sebmask = (M_downlw.index > sdate) & (M_downlw.index < edate)
fns = glob('/Volumes/seagate_desktop/idealized/case3/000101/wrfo*')
wrflist = list()
for fn in fns:
wrflist.append(Dataset(fn))
#cldfra = getvar(wrflist, "CLDFRA", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
z = getvar(wrflist, "z").mean('south_north').mean('west_east')
#cldfra_df = pd.DataFrame(cldfra.values, index = cldfra.Time.values, columns = z)
lh = getvar(wrflist, "LH", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lh_df = pd.DataFrame(lh.values, index = lh.Time.values)
sh = getvar(wrflist, "HFX", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
sh_df = pd.DataFrame(sh.values, index = sh.Time.values)
lwdnb = getvar(wrflist, "LWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwdnb_df = pd.DataFrame(lwdnb.values, index = lwdnb.Time.values)
lwupb = getvar(wrflist, "LWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwupb_df = pd.DataFrame(lwupb.values, index = lwupb.Time.values)
swdnb = getvar(wrflist, "SWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swdnb_df = pd.DataFrame(swdnb.values, index = swdnb.Time.values)
swupb = getvar(wrflist, "SWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swupb_df = pd.DataFrame(swupb.values, index = swupb.Time.values)
# MODIFIED
# Finding all wrfout file
fns = glob('/Volumes/seagate_desktop/idealized/landusetbl_modifications/3ClearSpring_000101/wrfo*')
# Creating an empty list to append to
wrflist = list()
# Opening the wrfout files and appending them to the empty list
for fn in fns:
wrflist.append(Dataset(fn))
# Importing WRF variables
## dbZ - for cloud plotting
cldfra_mod = getvar(wrflist, "CLDFRA", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
z_mod = getvar(wrflist, "z").mean('south_north').mean('west_east')
cldfra_df_mod = pd.DataFrame(cldfra_mod.values, index = cldfra_mod.Time.values, columns = z_mod)
## lh - latent heat flux
lh_mod = getvar(wrflist, "LH", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lh_df_mod = pd.DataFrame(lh_mod.values, index = lh_mod.Time.values)
## sh - sensible heat flux
sh_mod = getvar(wrflist, "HFX", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
sh_df_mod = pd.DataFrame(sh_mod.values, index = sh_mod.Time.values)
## lwdnb - downwelling longwave radiation
lwdnb_mod = getvar(wrflist, "LWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwdnb_df_mod = pd.DataFrame(lwdnb_mod.values, index = lwdnb_mod.Time.values)
## lwupb - upwelling longwave radiation
lwupb_mod = getvar(wrflist, "LWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
lwupb_df_mod = pd.DataFrame(lwupb_mod.values, index = lwupb_mod.Time.values)
## swdnb - downwelling shortwave radiation
swdnb_mod = getvar(wrflist, "SWDNB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swdnb_df_mod = pd.DataFrame(swdnb_mod.values, index = swdnb_mod.Time.values)
## swupb - upwelling shortwave radiation
swupb_mod = getvar(wrflist, "SWUPB", timeidx=ALL_TIMES, method="cat").mean('south_north').mean('west_east')
swupb_df_mod = pd.DataFrame(swupb_mod.values, index = swupb_mod.Time.values)
wrfstat = xr.open_dataset('/Volumes/seagate_desktop/idealized/case3/000101/second_try/wrfstat_d01_2015-05-22_00:00:00')
cst_qc = wrfstat['CSP_QC']
cst_tsk = wrfstat['CST_TSK']
cst_sh = wrfstat['CST_SH']
cst_lh = wrfstat['CST_LH']
cst_time = wrfstat['Times']
csp_z = wrfstat['CSP_Z']
csp_u = wrfstat['CSP_U']
csp_v = wrfstat['CSP_V']
csv_qv = wrfstat["CSP_QV"]
qc = pd.DataFrame(cst_qc.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
sh_df = pd.DataFrame(cst_sh.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'), columns = ['sh'])
lh_df = pd.DataFrame(cst_lh.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'), columns = ['lh'])
tsk_df = pd.DataFrame(cst_tsk.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'), columns = ['tsk'])
u_df = pd.DataFrame(csp_u.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
v_df = pd.DataFrame(csp_v.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
ws_df = np.sqrt(v_df**2 + u_df**2)
qv_df = pd.DataFrame(csv_qv.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
cloudmask = wrfstat['CSV_CLDFRAC'].isel(south_north = round(len(wrfstat.south_north) / 2), west_east = round(len(wrfstat.west_east) / 2))
cloudmask_df = pd.DataFrame(cloudmask.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
wrfstat = xr.open_dataset('/Volumes/seagate_desktop/idealized/landusetbl_modifications/3ClearSpring_000101/corrected_input/wrfstat_d01_2015-05-22_00:00:00')
cst_qc = wrfstat['CSP_QC']
cst_tsk = wrfstat['CST_TSK']
cst_sh = wrfstat['CST_SH']
cst_lh = wrfstat['CST_LH']
cst_time = wrfstat['Times']
csp_z = wrfstat['CSP_Z']
csp_u = wrfstat['CSP_U']
csp_v = wrfstat['CSP_V']
csv_qv = wrfstat["CSP_QV"]
qc_mod = pd.DataFrame(cst_qc.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
sh_df_mod = pd.DataFrame(cst_sh.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'), columns = ['sh'])
lh_df_mod = pd.DataFrame(cst_lh.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'), columns = ['lh'])
tsk_df_mod = pd.DataFrame(cst_tsk.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'), columns = ['tsk'])
u_df_mod = pd.DataFrame(csp_u.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
v_df_mod = pd.DataFrame(csp_v.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
ws_df_mod = np.sqrt(v_df_mod**2 + u_df_mod**2)
qv_df_mod = pd.DataFrame(csv_qv.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
cloudmask = wrfstat['CSV_CLDFRAC'].isel(south_north = round(len(wrfstat.south_north) / 2), west_east = round(len(wrfstat.west_east) / 2))
cloudmask_df_mod = pd.DataFrame(cloudmask.values,
index = pd.date_range(start='5/22/2015 00:00:00', end='5/24/2015 22:30:00', freq = '30min'),
columns = csp_z.isel(Time = 1).values)
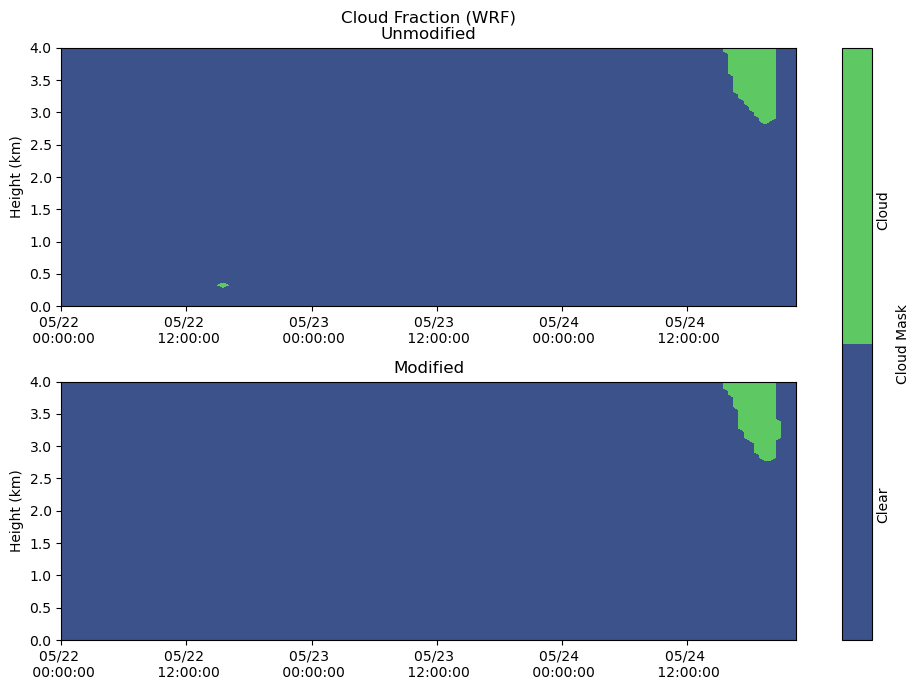
Clouds¶
fig, axs = plt.subplots(2, figsize=(10,7))
heatmap = axs[0].contourf(cloudmask_df.index, cloudmask_df.columns / 1000, cloudmask_df.T.values, levels = [-1, 0, 1])
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Cloud Fraction (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,4)
heatmap = axs[1].contourf(cloudmask_df_mod.index, cloudmask_df_mod.columns / 1000, cloudmask_df_mod.T, levels = [-1, 0, 1])
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,4)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[:], ticks = [-0.5, 0.5], label = "Cloud Mask")
cbar.ax.set_yticklabels(['Clear','Cloud'], rotation = 90)
cbar.ax.tick_params(size=0)
plt.show()
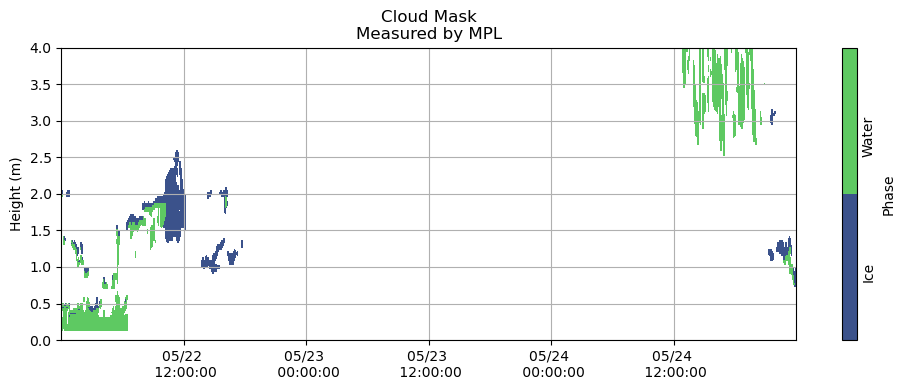
fns = ['/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150522.cdf',
'/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150523.cdf',
'/Volumes/seagate_desktop/data/MPL/Robert_MPLData/FinalNICELidarData/NICE_MPLDataFinal20150524.cdf']
measured_cloudmask_all = pd.DataFrame()
for fn in fns:
rbt_cldmask = xr.open_dataset(fn)
# Time is in UTC time stamp in fractional hours
Time = rbt_cldmask.variables['DataTime'].values
# Fix time to be a timestamp - Currently in hours
yy = 2015
mm = int(fn[-8:-6])
dd = int(fn[-6:-4])
date = [datetime(yy,mm,dd) + timedelta(seconds=hr*3600) for hr in Time[~np.isnan(Time)]]
measured_cloudmask = pd.DataFrame(rbt_cldmask['PhaseMask'][~np.isnan(Time)].values, index = date, columns = rbt_cldmask['Range'])
measured_cloudmask_all = pd.concat([measured_cloudmask_all, measured_cloudmask])
fig, ax = plt.subplots(figsize=(10,4))
plt.grid()
heatmap = plt.contourf(measured_cloudmask_all.index,
measured_cloudmask_all.columns / 1000,
measured_cloudmask_all.T.values,
[0, 1, 2])
# nan - no information
# 1 - cloud liquid
# 2 - cloud ice
plt.title('Cloud Mask\nMeasured by MPL')
plt.ylabel('Height (m)')
cbar = fig.colorbar(heatmap, label = 'Phase', ticks = [0.5, 1.5], orientation = 'vertical')
cbar.ax.set_yticklabels(['Ice', 'Water'], rotation = 90)
cbar.ax.tick_params(size=0)
plt.gca().xaxis.set_major_formatter(myFmt)
plt.ylim(0, 4)
plt.tight_layout()
plt.show()
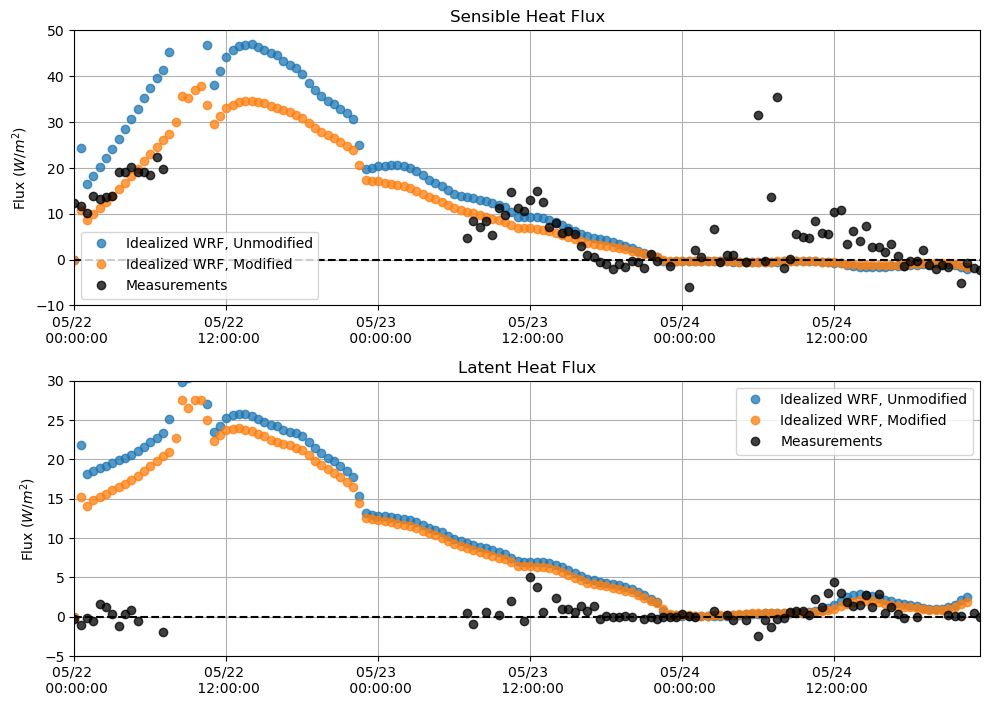
Sensible and Latent Heat Flux¶
plt.figure(figsize = (10,7))
plt.subplot(211)
plt.plot(sh_df, 'o', alpha = 0.75)
plt.plot(sh_df_mod, 'o', alpha = 0.75)
plt.plot(M_sen[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.hlines(0, xmin = M_lat[sdate:edate].index[0], xmax = M_lat[sdate:edate].index[-1], linestyle = '--', color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Sensible Heat Flux')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(-10,50)
plt.xlim(M_lat[sdate:edate].index[0],M_lat[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(212)
plt.plot(lh_df, 'o', alpha = 0.75)
plt.plot(lh_df_mod, 'o', alpha = 0.75)
plt.plot(M_lat[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.hlines(0, xmin = M_lat[sdate:edate].index[0], xmax = M_lat[sdate:edate].index[-1], linestyle = '--', color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Latent Heat Flux')
plt.grid()
plt.ylim(-5,30)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_lat[sdate:edate].index[0],M_lat[sdate:edate].index[-1])
plt.tight_layout()
plt.gca().xaxis.set_major_formatter(myFmt)
plt.show()
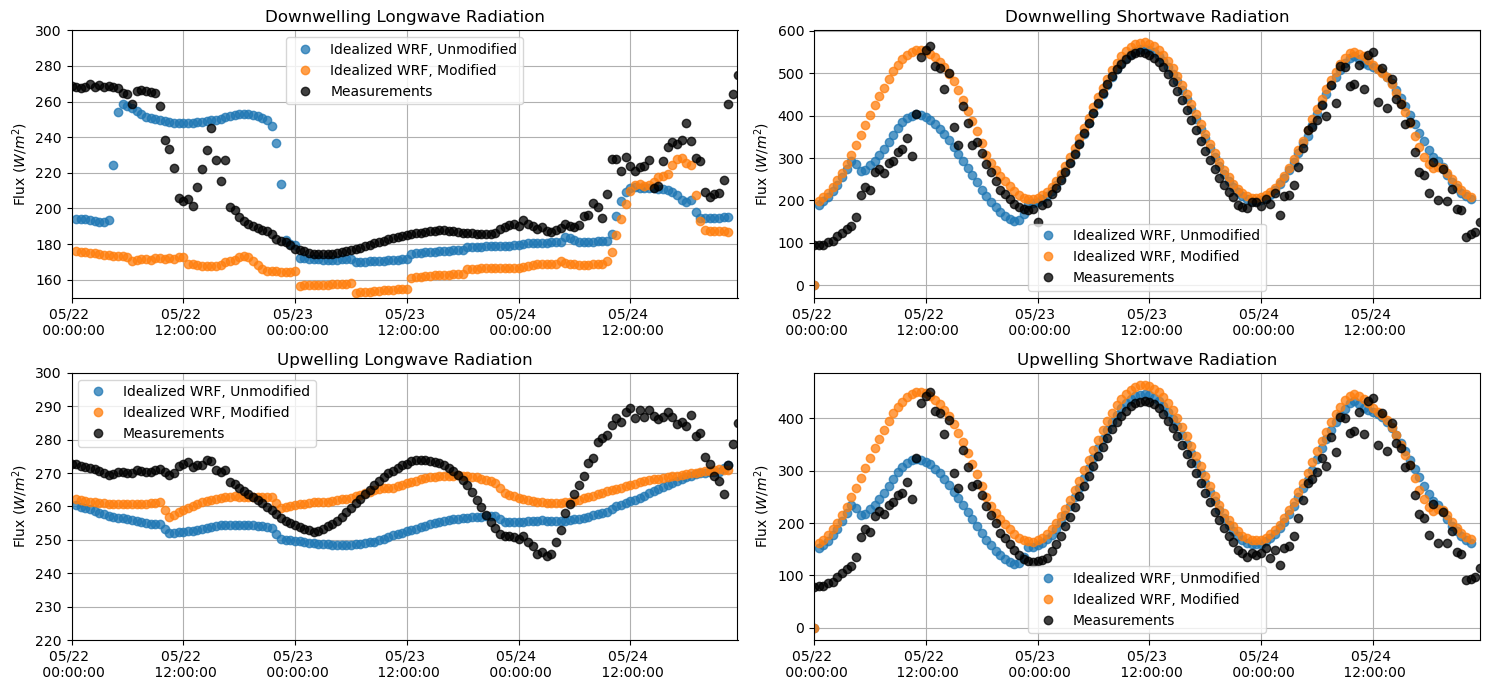
Longwave and Shortwave Radiation¶
plt.figure(figsize = (15,7))
plt.subplot(221)
plt.plot(lwdnb_df, 'o', alpha = 0.75)
plt.plot(lwdnb_df_mod, 'o', alpha = 0.75)
plt.plot(M_downlw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Downwelling Longwave Radiation')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(150,300)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(223)
plt.plot(lwupb_df, 'o', alpha = 0.75)
plt.plot(lwupb_df_mod, 'o', alpha = 0.75)
plt.plot(M_uplw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Upwelling Longwave Radiation')
plt.grid()
plt.ylim(220,300)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(222)
plt.plot(swdnb_df, 'o', alpha = 0.75)
plt.plot(swdnb_df_mod, 'o', alpha = 0.75)
plt.plot(M_downsw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Downwelling Shortwave Radiation')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
#plt.ylim(180,300)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.subplot(224)
plt.plot(swupb_df, 'o', alpha = 0.75)
plt.plot(swupb_df_mod, 'o', alpha = 0.75)
plt.plot(M_upsw[sdate:edate], 'o', alpha = 0.75, color = 'k')
plt.ylabel('Flux $(W/m^{2})$')
plt.title('Upwelling Shortwave Radiation')
plt.grid()
#plt.ylim(180, 300)
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.tight_layout()
plt.show()
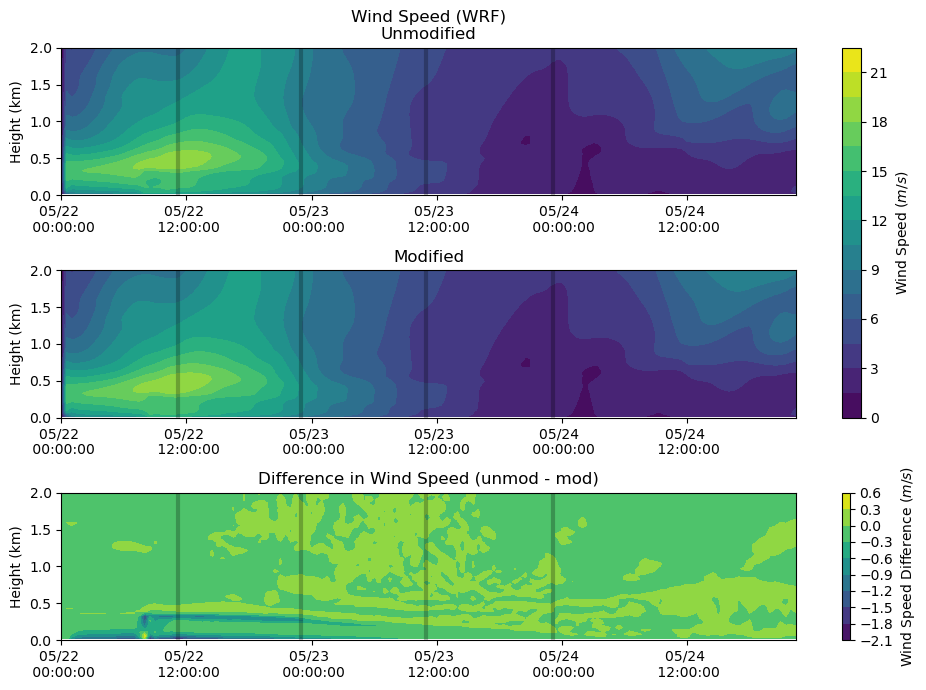
Wind Speed¶
fig, axs = plt.subplots(3, figsize=(10,7))
heatmap = axs[0].contourf(ws_df.index, ws_df.columns / 1000, ws_df.T.values, levels = 20)
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Wind Speed (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,2)
axs[0].vlines(sounding_ws[sdate:'2015-05-24'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap = axs[1].contourf(ws_df_mod.index, ws_df_mod.columns / 1000, ws_df_mod.T.values, levels = 20)
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,2)
axs[1].vlines(sounding_ws[sdate:'2015-05-24'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap_1 = axs[2].contourf(ws_df.index, ws_df.columns / 1000, ws_df.T.values - ws_df_mod.T.values)
axs[2].set_ylabel('Height (km)')
axs[2].set_title('Difference in Wind Speed (unmod - mod)')
axs[2].xaxis.set_major_formatter(myFmt)
axs[2].set_ylim(0,2)
axs[2].vlines(sounding_ws[sdate:'2015-05-24'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[0:2])
cbar.set_label('Wind Speed ($m/s$)')
cbar = fig.colorbar(heatmap_1, ax=axs[2])
cbar.set_label('Wind Speed Difference ($m/s$)')
plt.show()
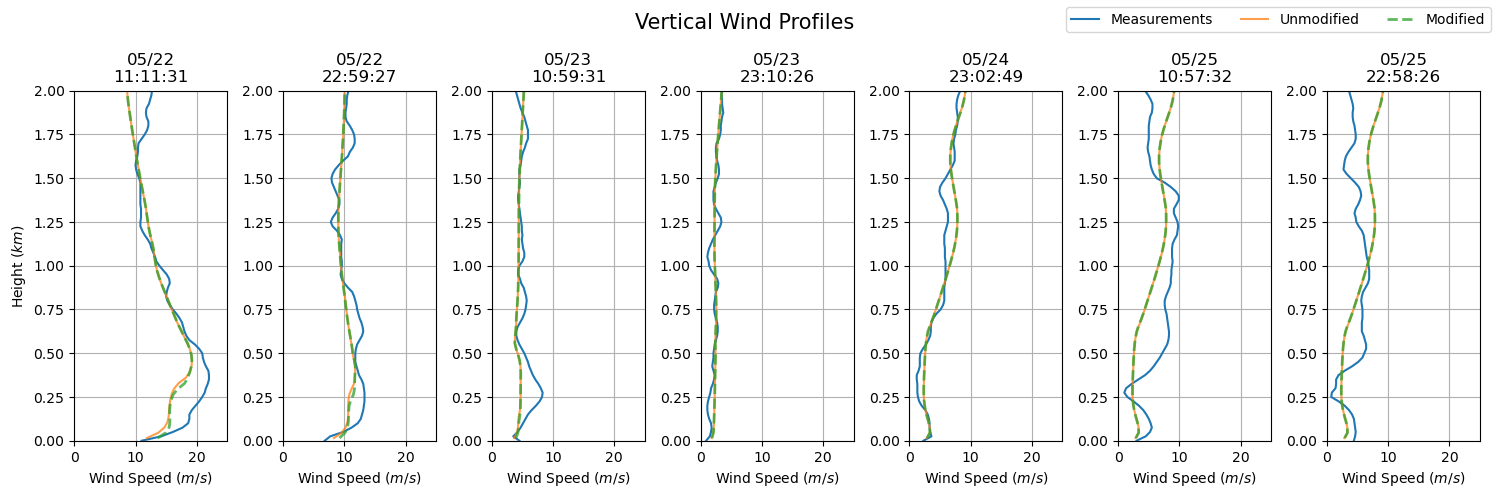
dates = sounding_ws[sdate:'2015-05-25'].index
fig, axs = plt.subplots(ncols = len(dates), figsize=(15,5))
for i in np.arange(0, len(dates), 1):
prof = axs[i].plot(sounding_ws.loc[dates[i]], sounding_ws.loc[dates[i]].index / 1000, label = 'Measurements')
axs[i].set_ylim(0, 2)
axs[i].grid()
axs[i].set_xlim(0, 25)
axs[i].set_xlabel('Wind Speed ($m/s$)')
axs[i].set_title(dates[i].strftime('%m/%d') + '\n' + dates[i].strftime('%H:%M:%S'))
axs[i].plot(ws_df.iloc[ws_df.index.get_loc(dates[i], method = 'nearest')],
ws_df.iloc[ws_df.index.get_loc(dates[0], method = 'nearest')].index / 1000,
label = 'Unmodified', alpha = 0.75)
axs[i].plot(ws_df_mod.iloc[ws_df_mod.index.get_loc(dates[i], method = 'nearest')],
ws_df_mod.iloc[ws_df_mod.index.get_loc(dates[0], method = 'nearest')].index / 1000,
'--', label = 'Modified', alpha = 0.75, lw = 2)
axs[0].set_ylabel('Height ($km$)')
plt.suptitle('Vertical Wind Profiles', fontsize = 15)
plt.tight_layout()
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right', ncol = 3)
plt.show()
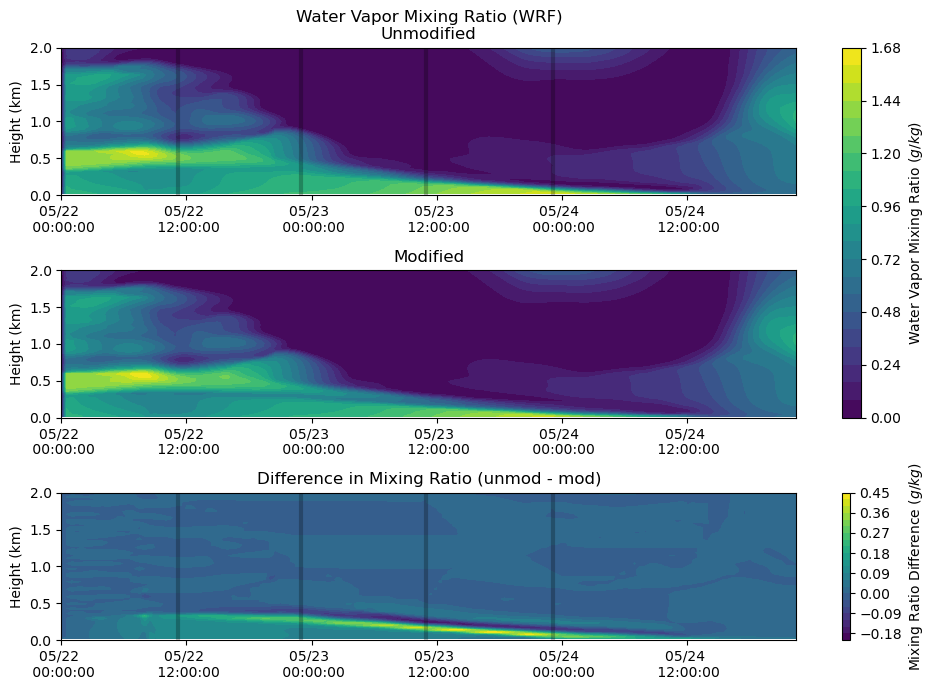
Moisture¶
fig, axs = plt.subplots(3, figsize=(10,7))
heatmap = axs[0].contourf(qv_df_mod.index, qv_df.columns / 1000, qv_df.T.values * 1000, levels = 20)
axs[0].set_ylabel('Height (km)')
axs[0].set_title('Water Vapor Mixing Ratio (WRF)\nUnmodified')
axs[0].xaxis.set_major_formatter(myFmt)
axs[0].set_ylim(0,2)
axs[0].vlines(sounding_ws[sdate:'2015-05-24'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap = axs[1].contourf(qv_df_mod.index, qv_df_mod.columns / 1000, qv_df_mod.T.values * 1000, levels = 20)
axs[1].set_ylabel('Height (km)')
axs[1].set_title('Modified')
axs[1].xaxis.set_major_formatter(myFmt)
axs[1].set_ylim(0,2)
axs[1].vlines(sounding_ws[sdate:'2015-02-24'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
heatmap_1 = axs[2].contourf(qv_df_mod.index, qv_df.columns / 1000,
(qv_df.T.values * 1000) - (qv_df_mod.T.values * 1000), levels = 20)
axs[2].set_ylabel('Height (km)')
axs[2].set_title('Difference in Mixing Ratio (unmod - mod)')
axs[2].xaxis.set_major_formatter(myFmt)
axs[2].set_ylim(0,2)
axs[2].vlines(sounding_ws[sdate:'2015-05-24'].index, 0, 2, color = 'k', alpha = 0.25, lw = 3)
plt.tight_layout()
cbar = fig.colorbar(heatmap, ax=axs[0:2])
cbar.set_label('Water Vapor Mixing Ratio ($g/kg$)')
cbar = fig.colorbar(heatmap_1, ax=axs[2])
cbar.set_label('Mixing Ratio Difference ($g/kg$)')
plt.show()
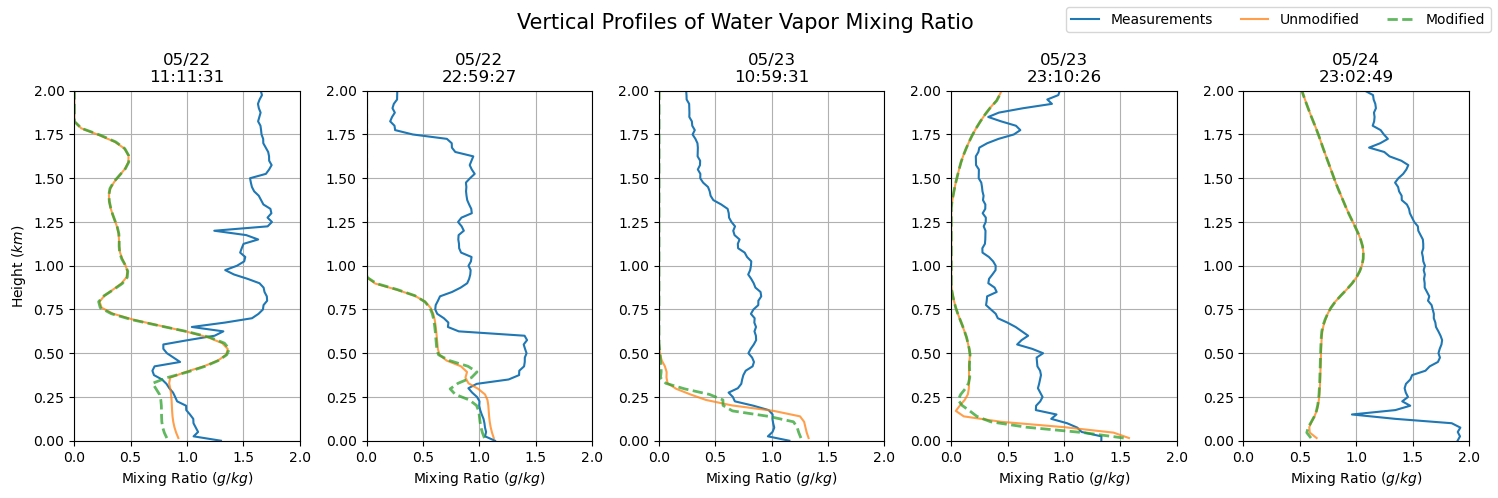
dates = sfc_mixing_ratio[sdate:'2015-05-24'].index
fig, axs = plt.subplots(ncols = len(dates), figsize=(15,5))
for i in np.arange(0, len(dates), 1):
prof = axs[i].plot(sfc_mixing_ratio.loc[dates[i].to_pydatetime()],
sfc_mixing_ratio.loc[dates[i].to_pydatetime()].index.astype(float) / 1000,
label = 'Measurements')
axs[i].set_ylim(0, 2)
axs[i].grid()
axs[i].set_xlim(0, 2)
axs[i].set_xlabel('Mixing Ratio ($g/kg$)')
axs[i].set_title(dates[i].strftime('%m/%d') + '\n' + dates[i].strftime('%H:%M:%S'))
axs[i].plot(qv_df.iloc[qv_df.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')] * 1000,
qv_df.iloc[qv_df.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')].index / 1000,
label = 'Unmodified', alpha = 0.75)
axs[i].plot(qv_df_mod.iloc[qv_df_mod.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')] * 1000,
qv_df_mod.iloc[qv_df_mod.index.get_loc(dates[i].to_pydatetime(), method = 'nearest')].index / 1000,
'--', label = 'Modified', alpha = 0.75, lw = 2)
axs[0].set_ylabel('Height ($km$)')
plt.suptitle('Vertical Profiles of Water Vapor Mixing Ratio', fontsize = 15)
plt.tight_layout()
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, loc='upper right', ncol = 3)
plt.show()
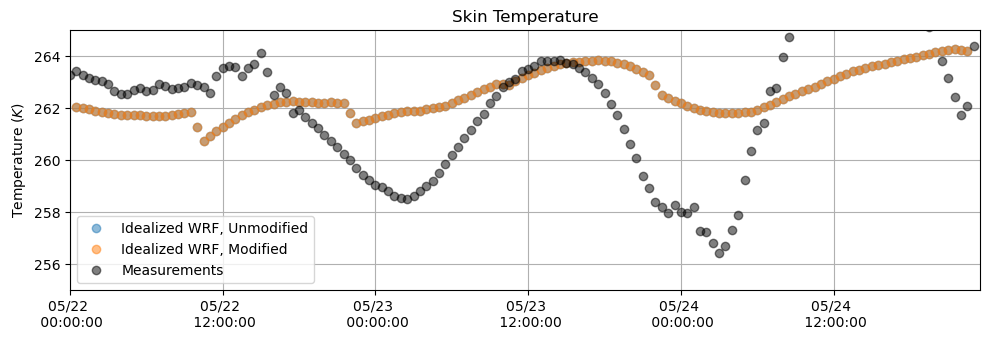
Temperature¶
plt.figure(figsize = (10,3.5))
plt.plot(tsk_df, 'o', alpha = 0.5)
plt.plot(tsk_df_mod, 'o', alpha = 0.5)
plt.plot(T_meas[sdate:edate].resample('30min').first(), 'o', alpha = 0.5, color = 'k')
plt.ylabel('Temperature $(K)$')
plt.title('Skin Temperature')
plt.grid()
plt.legend(['Idealized WRF, Unmodified', 'Idealized WRF, Modified', 'Measurements'])
plt.ylim(255,265)
plt.xlim(M_downlw[sdate:edate].index[0],M_downlw[sdate:edate].index[-1])
plt.gca().xaxis.set_major_formatter(myFmt)
plt.tight_layout()
plt.show()
Summary Table¶
Red highlighting indicates the lowest correlation of that variable, green indicates the highest.
edate = '2015-05-24 22:30:00'
lwdns = M_downlw[sdate:edate]
lwdns.columns = ['measured']
lwdns['unmodified'] = lwdnb_df[sdate:edate]
lwdns['modified'] = lwdnb_df_mod[sdate:edate]
lwups = M_uplw[sdate:edate]
lwups.columns = ['measured']
lwups['unmodified'] = lwupb_df[sdate:edate]
lwups['modified'] = lwupb_df_mod[sdate:edate]
swdns = M_downsw[sdate:edate]
swdns.columns = ['measured']
swdns['unmodified'] = swdnb_df[sdate:edate]
swdns['modified'] = swdnb_df_mod[sdate:edate]
swups = M_upsw[sdate:edate]
swups.columns = ['measured']
swups['unmodified'] = swupb_df[sdate:edate]
swups['modified'] = swupb_df_mod[sdate:edate]
lhs = M_lat[sdate:edate]
lhs.columns = ['measured']
lhs['unmodified'] = lh_df[sdate:edate]
lhs['modified'] = lh_df_mod[sdate:edate]
shs = M_sen[sdate:edate]
shs.columns = ['measured']
shs['unmodified'] = sh_df[sdate:edate]
shs['modified'] = sh_df_mod[sdate:edate]
correlation_coefficients = pd.DataFrame([lhs.corr()['measured'].values,
shs.corr()['measured'].values,
lwdns.corr()['measured'].values,
lwups.corr()['measured'].values,
swdns.corr()['measured'].values,
swups.corr()['measured'].values],
columns = lhs.corr().columns,
index = ['Latent', 'Sensible', 'Downwelling Longwave', 'Upwelling Longwave', 'Downwelling Shortwave', 'Upwelling Shortwave']).T
cc_3 = correlation_coefficients
r_squared = correlation_coefficients ** 2
rs_3 = r_squared
r_squared[1:].style.highlight_max(color = 'lightgreen', axis = 0).highlight_min(color = 'pink', axis = 0)
| Latent | Sensible | Downwelling Longwave | Upwelling Longwave | Downwelling Shortwave | Upwelling Shortwave | |
|---|---|---|---|---|---|---|
| unmodified | 0.019199 | 0.399344 | 0.142917 | 0.011557 | 0.801807 | 0.803250 |
| modified | 0.018927 | 0.390447 | 0.070061 | 0.001145 | 0.821090 | 0.840806 |
Statistics and Summary¶
mean_cc = (cc_3.iloc[:,0:4] + cc_2.iloc[:,0:4] + cc_1)/3
mean_cc_sw = ((cc_3.iloc[:,4:6] + cc_2.iloc[:,4:6])/2)
mean_cc = pd.concat([mean_cc, mean_cc_sw])
mean_rs = (rs_3.iloc[:,0:4] + rs_2.iloc[:,0:4] + rs_1)/3
mean_rs_sw = ((rs_3.iloc[:,4:6] + rs_2.iloc[:,4:6])/2)
mean_rs = pd.concat([mean_rs, mean_rs_sw], axis=1, join="inner")
mean_rs[1:].style.highlight_max(color = 'lightgreen', axis = 0).highlight_min(color = 'pink', axis = 0)
| Latent | Sensible | Downwelling Longwave | Upwelling Longwave | Downwelling Shortwave | Upwelling Shortwave | |
|---|---|---|---|---|---|---|
| unmodified | 0.007632 | 0.145286 | 0.315652 | 0.209108 | 0.843545 | 0.843282 |
| modified | 0.009108 | 0.147682 | 0.307759 | 0.205637 | 0.853632 | 0.868321 |